HISTOGRAM Statement: CAPABILITY Procedure
Example 5.14 Annotating a Folded Normal Curve
See FNORM2 in the SAS/QC Sample LibraryThis example shows how to display a fitted curve that is not supported by the HISTOGRAM statement.
The offset of an attachment point is measured (in mm) for a number of manufactured assemblies, and the measurements are saved
in a data set named Assembly.
data Assembly; label Offset = 'Offset (in mm)'; input Offset @@; datalines; 11.11 13.07 11.42 3.92 11.08 5.40 11.22 14.69 6.27 9.76 9.18 5.07 3.51 16.65 14.10 9.69 16.61 5.67 2.89 8.13 9.97 3.28 13.03 13.78 3.13 9.53 4.58 7.94 13.51 11.43 11.98 3.90 7.67 4.32 12.69 6.17 11.48 2.82 20.42 1.01 3.18 6.02 6.63 1.72 2.42 11.32 16.49 1.22 9.13 3.34 1.29 1.70 0.65 2.62 2.04 11.08 18.85 11.94 8.34 2.07 0.31 8.91 13.62 14.94 4.83 16.84 7.09 3.37 0.49 15.19 5.16 4.14 1.92 12.70 1.97 2.10 9.38 3.18 4.18 7.22 15.84 10.85 2.35 1.93 9.19 1.39 11.40 12.20 16.07 9.23 0.05 2.15 1.95 4.39 0.48 10.16 4.81 8.28 5.68 22.81 0.23 0.38 12.71 0.06 10.11 18.38 5.53 9.36 9.32 3.63 12.93 10.39 2.05 15.49 8.12 9.52 7.77 10.70 6.37 1.91 8.60 22.22 1.74 5.84 12.90 13.06 5.08 2.09 6.41 1.40 15.60 2.36 3.97 6.17 0.62 8.56 9.36 10.19 7.16 2.37 12.91 0.95 0.89 3.82 7.86 5.33 12.92 2.64 7.92 14.06 ;
The assembly process is in statistical control, and it is decided to fit a folded normal distribution to the offset measurements. A variable X has a folded normal distribution if ![]() , where Y is distributed as
, where Y is distributed as ![]() . The fitted density is
. The fitted density is
|
|
You can use SAS/IML software to compute preliminary estimates of ![]() and
and ![]() based on a method of moments given by Elandt (1961). These estimates are computed by solving equation (19) of Elandt (1961), which is given by
based on a method of moments given by Elandt (1961). These estimates are computed by solving equation (19) of Elandt (1961), which is given by
|
|
where ![]() is the standard normal distribution function, and
is the standard normal distribution function, and
|
|
Then the estimates of ![]() and
and ![]() are given by
are given by
![\[ \begin{array}{lll} \hat{\sigma }_0 & = \sqrt { \frac{ \frac{1}{n} \sum _{i=1}^ n x_ i^2}{1 + \hat{\theta }^2 } } \\[.1in] \hat{\mu }_0 & = \hat{\theta } \cdot \hat{\sigma }_0 \end{array} \]](images/qcug_capability0517.png) |
Begin by using the MEANS procedure to compute the first and second moments and using the DATA step to compute the constant A.
proc means data = Assembly noprint; var Offset; output out=Stat mean=m1 var=var n=n min = min; run; * Compute constant A from equation (19) of Elandt (1961) ; data Stat; keep m2 a min; set Stat; a = (m1*m1); m2 = ((n-1)/n)*var + a; a = a/m2; run;
Next, use the SAS/IML subroutine NLPDD to solve equation (19) by minimizing ![]() , and compute
, and compute ![]() and
and ![]() .
.
proc iml;
use Stat;
read all var {m2} into m2;
read all var {a} into a;
read all var {min} into min;
* f(t) is the function in equation (19) of Elandt (1961) ;
start f(t) global(a);
y = .39894*exp(-0.5*t*t);
y = (2*y-(t*(1-2*probnorm(t))))**2/(1+t*t);
y = (y-a)**2;
return(y);
finish;
* Minimize (f(t)-A)**2 and estimate mu and sigma ;
if ( min < 0 ) then do;
print "Warning: Observations are not all nonnegative.";
print " The folded normal is inappropriate.";
stop;
end;
if ( a < 0.637 ) then do;
print "Warning: the folded normal may be inappropriate";
end;
opt = { 0 0 };
con = { 1e-6 };
x0 = { 2.0 };
tc = { . . . . . 1e-12 . . . . . . .};
call nlpdd(rc,etheta0,"f",x0,opt,con,tc);
esig0 = sqrt(m2/(1+etheta0*etheta0));
emu0 = etheta0*esig0;
create Prelim var {emu0 esig0 etheta0};
append;
close Prelim;
* Define the log likelihood of the folded normal ;
start g(p) global(x);
y = 0.0;
do i = 1 to nrow(x);
z = exp( (-0.5/p[2])*(x[i]-p[1])*(x[i]-p[1]) );
z = z + exp( (-0.5/p[2])*(x[i]+p[1])*(x[i]+p[1]) );
y = y + log(z);
end;
y = y - nrow(x)*log( sqrt( p[2] ) );
return(y);
finish;
* Maximize the log likelihood with subroutine NLPDD ;
use Assembly;
read all var {Offset} into x;
esig0sq = esig0*esig0;
x0 = emu0||esig0sq;
opt = { 1 0 };
con = { . 0.0, . . };
call nlpdd(rc,xr,"g",x0,opt,con);
emu = xr[1];
esig = sqrt(xr[2]);
etheta = emu/esig;
create Parmest var{emu esig etheta};
append;
close Parmest;
quit;
title 'The Data Set Prelim'; proc print data=Prelim noobs; run;
The preliminary estimates are saved in the data set Prelim, as shown in Output 5.14.1.
Output 5.14.1: Preliminary Estimates of ![]() ,
, ![]() , and
, and ![]()
| The Data Set Prelim |
| EMU0 | ESIG0 | ETHETA0 |
|---|---|---|
| 6.51735 | 6.54953 | 0.99509 |
Now, using ![]() and
and ![]() as initial estimates, call the NLPDD subroutine to maximize the log likelihood,
as initial estimates, call the NLPDD subroutine to maximize the log likelihood, ![]() , of the folded normal distribution, where, up to a constant,
, of the folded normal distribution, where, up to a constant,
|
|
* Define the log likelihood of the folded normal ;
start g(p) global(x);
y = 0.0;
do i = 1 to nrow(x);
z = exp( (-0.5/p[2])*(x[i]-p[1])*(x[i]-p[1]) );
z = z + exp( (-0.5/p[2])*(x[i]+p[1])*(x[i]+p[1]) );
y = y + log(z);
end;
y = y - nrow(x)*log( sqrt( p[2] ) );
return(y);
finish;
* Maximize the log likelihood with subroutine NLPDD ;
use assembly;
read all var {offset} into x;
esig0sq = esig0*esig0;
x0 = emu0||esig0sq;
opt = { 1 0 };
con = { . 0.0, . . };
call nlpdd(rc,xr,"g",x0,opt,con);
emu = xr[1];
esig = sqrt(xr[2]);
etheta = emu/esig;
create parmest var{emu esig etheta};
append;
close parmest;
quit;
title 'The Data Set PARMEST'; proc print data=Parmest noobs; var emu esig etheta; run;
The data set Parmest saves the maximum likelihood estimates ![]() and
and ![]() (as well as
(as well as ![]() ), as shown in Output 5.14.2.
), as shown in Output 5.14.2.
Output 5.14.2: Final Estimates of ![]() ,
, ![]() , and
, and ![]()
| The Data Set PARMEST |
| EMU | ESIG | ETHETA |
|---|---|---|
| 6.66761 | 6.39650 | 1.04239 |
To annotate the curve on a histogram, begin by computing the width and endpoints of the histogram intervals. The following statements save these values in an OUTFIT= data set called OUT. Note that a plot is not produced at this point.
ods graphics off; proc capability data = Assembly noprint; histogram Offset / outfit = Out normal(noprint) noplot; run;
title 'OUTFIT= Data Set Out';
proc print data=Out noobs round;
var _var_ _curve_ _locatn_ _scale_ _chisq_ _df_ _pchisq_
_midpt1_ _width_ _midptn_ _expect_ _eststd_ _adasq_
_adp_ _cvmwsq_ _cvmp_ _ksd_ _ksp_;
run;
Output 5.14.3 provides a partial listing of the data set Out. The width and endpoints of the histogram bars are saved as values of the variables _WIDTH_, _MIDPT1_, and _MIDPTN_. See Output Data Sets.
Output 5.14.3: The OUTFIT= Data Set Out
| OUTFIT= Data Set Out |
| _VAR_ | _CURVE_ | _LOCATN_ | _SCALE_ | _CHISQ_ | _DF_ | _PCHISQ_ | _MIDPT1_ | _WIDTH_ | _MIDPTN_ | _EXPECT_ | _ESTSTD_ | _ADASQ_ | _ADP_ | _CVMWSQ_ | _CVMP_ | _KSD_ | _KSP_ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Offset | NORMAL | 7.62 | 5.24 | 31.17 | 5 | 0 | 1.5 | 3 | 22.5 | 7.62 | 5.24 | 1.9 | 0.01 | 0.28 | 0.01 | 0.09 | 0.01 |
The following statements create an annotate data set named Anno, which contains the coordinates of the fitted curve:
data Anno;
merge Parmest Out;
length function color $ 8;
function = 'point';
color = 'black';
size = 2;
xsys = '2';
ysys = '2';
when = 'a';
constant = 39.894*_width_;;
left = _midpt1_ - .5*_width_;
right = _midptn_ + .5*_width_;
inc = (right-left)/100;
do x = left to right by inc;
z1 = (x-emu)/esig;
z2 = (x+emu)/esig;
y = (constant/esig)*(exp(-0.5*z1*z1)+exp(-0.5*z2*z2));
output;
function = 'draw';
end;
run;
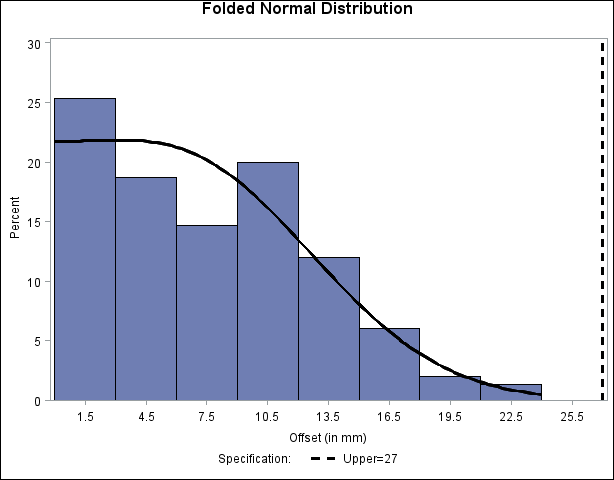
The following statements read the ANNOTATE= data set and display the histogram and fitted curve, as shown in Output 5.14.4:
ods graphics off; title "Folded Normal Distribution"; proc capability data=Assembly noprint; spec usl=27 cusl=black lusl=2 wusl=2; histogram Offset / annotate = Anno; run;
Output 5.14.4: Histogram with Annotated Folded Normal Curve