| PROC MVPMODEL Statement |
- PROC MVPMODEL <options> ;
Option |
Description |
|---|---|
Computes the principal components from the covariance matrix |
|
Performs tcross validation to select the number of principal components |
|
Specifies the input data set |
|
Specifies how observations with missing values are handled |
|
Specifies the number of principal components to extract |
|
Suppresses centering of process variables before fitting the model |
|
Suppresses re-centering and rescaling of process variables before each model is fit in the cross validation |
|
Suppresses the display of all output |
|
Suppresses scaling of process variables before fitting the model |
|
Specifies the output data set |
|
Specifies the output data set for loadings (eigenvectors) |
|
Requests and specifies details of plots |
|
Specifies the prefix for naming principal component score variables in the OUT= data set |
|
Specifies the prefix for naming residual variables in OUT= data set |
|
Standardizes the principal component scores |
|
Specifies the variable in the input data set used to group the data when there are multiple observations per time value |
The following list provides details about these options.
- COV
- NOSCALE
-
computes the principal components from the covariance matrix. By default, the correlation matrix is analyzed. Use of the COV option causes variables with large variances to be more strongly associated with components with large eigenvalues and causes variables with small variances to be more strongly associated with components with small eigenvalues. You should not specify the COV option unless the units in which the variables are measured are comparable or the variables are standardized in some way.
Specifying the NOSCALE option is equivalent to specifying the COV option.
- CV=ONE
- CV=SPLIT <(n)>
- CV=BLOCK <(n)>
- CV=RANDOM <(cv-random-options)>
-
specifies the cross validation method to be used. By default, no cross validation is performed. The option CV=ONE requests one-at-a-time cross validation, CV=SPLIT requests that every nth observation be excluded, CV=BLOCK requests that n blocks of consecutive observations be excluded, and CV=RANDOM requests that observations be excluded at random. Optionally, you can specify n for CV=SPLIT and CV=BLOCK; the default is n=7 in each case. You can also specify the following optional cv-random-options in parentheses after the CV=RANDOM option:
- NITER=n
specifies the number of random subsets to exclude. The default value is 10.
- NTEST=n
specifies the number of observations in each random subset chosen for exclusion. The default value is one-tenth of the total number of observations.
- SEED=n
specifies an integer used to start the pseudo-random number generator for selecting the random test set. If you do not specify a seed or specify a value less than or equal to zero, the seed is generated by default from reading the time of day from the computer’s clock.
You cannot specify the CV= option together with the NCOMP= option. See the section Cross Validation for more information.
- DATA=SAS-data-set
specifies the input SAS data set to be analyzed. If the DATA= option is omitted, the procedure uses the most recently created SAS data set.
- MISSING=(AVG | NONE)
specifies how observations with missing values are to be handled in computing the fit. The default is MISSING=NONE, which excludes observations with missing values for any process variables from the analysis. MISSING=AVG specifies that the fit be computed by replacing missing values of a process variable with the average of its nonmissing values.
- NCOMP=n
specifies the number of principal components to extract. The default is
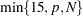 , where
, where 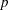 is the number of process variables and
is the number of process variables and 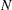 is the number of observations (runs). You cannot specify the NCOMP= option together with the CV= option.
is the number of observations (runs). You cannot specify the NCOMP= option together with the CV= option. - NOCENTER
suppresses centering of the process variables before fitting. This is useful if the variables are already centered and scaled. See the section Centering and Scaling for more information.
- NOCVSTDIZE
suppresses re-centering and rescaling of the process variables before each model is fit in the cross validation. See the section Centering and Scaling for more information.
- NOPRINT
suppresses the display of all results, both tabular and graphical. This is useful when you only want to produce output data sets.
- NOSCALE
- COV
-
suppresses scaling of the process variables before fitting. This is useful if the variables are already centered and scaled.
Specifying the COV option is equivalent to specifying the NOSCALE option.
- OUT=SAS-data-set
creates an output data set that contains all the original data from the input data set, principal component scores, and multivariate summary statistics. See the section Output Data Sets for details.
- OUTLOADINGS=SAS-data-set
creates an output data set that contains the loadings for the principal components and the eigenvalues of the correlation (or covariance) matrix. See the section Output Data Sets for details.
- PLOTS <(global-plot-options)> <= plot-request <(options)>>
- PLOTS <(global-plot-options)> <= (plot-request <(options)> <... plot-request <(options)>>)>
-
controls the plots produced through ODS Graphics. When you specify only one plot request, you can omit the parentheses from around the plot request. For example:
plots=none plots=score plots=loadings
ODS Graphics must be enabled before you request plots.
For general information about ODS Graphics, see Chapter 21, Statistical Graphics Using ODS (SAS/STAT User's Guide).
The global-plot-options include the following:
- FLIP
interchanges the X-axis and Y-axis dimensions for the score and loading plots.
- NCOMP=n
specifies that pairwise score or loading plots be produced for the first n principal components. The number of score or loading plots produced is
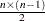 .
. - ONLY
suppresses the default plots. Only plots specifically requested are displayed. The default plots are the CV plot when the CV= option is specified, and the scree and variation-explained plots otherwise.
The plot-requests include the following:
- ALL
produces all appropriate plots.
- CVPLOT
produces a plot that displays results of the cross validation and R-square analysis. This plot requires the CV= option to be specified and is displayed by default in that case.
- LOADINGS <(NCOMP=n | FLIP)>
produces pairwise scatter plots of the principal component loadings. You can use NCOMP=n to specify the number of principal components for which plots are produced, and the FLIP option to interchange the default X-axis and Y-axis dimensions.
- NONE
suppresses the display of all plots.
- SCORES <(score-options)>
-
produces pairwise scatter plots of the principal component scores. You can use the NCOMP= option to control the number of plots that are displayed.
The available score-options are as follows:
- ALPHA=value
specifies the probability used to compute a prediction ellipse that is overlaid on the score plot. The default value is 0.05. If you specify the ALPHA= option, you do not need to specify the ELLIPSE option.
- ELLIPSE
requests that a prediction ellipse be overlaid on the principal component score plots. The probability that a new observation falls outside the prediction ellipse is specified by the ALPHA= option.
- FLIP
flips or interchanges the X-axis and Y-axis dimensions for the score plots. Specify PLOTS=SCORES(FLIP) to flip the X-axis and Y-axis dimensions.
- GROUP=variable
specifies a variable in the input data set used to group the points on the score plots. Points with different GROUP= variable values are plotted using different markers and colors to distinguish the groups.
- NCOMP=n
specifies that pairwise score plots be produced for the first n principal components. The default is 5 or the total number of components
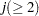 , whichever is smaller. If n
, whichever is smaller. If n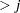 , NCOMP= j is used. Be aware that the number of plots (
, NCOMP= j is used. Be aware that the number of plots (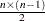 ) produced grows quadratically when n increases.
) produced grows quadratically when n increases. - NOLABELS
suppresses labels for the points on the score plots. By default, points are labeled with the values of the first variable listed in the ID statement, or the observation number if no ID statement is specified.
- SCREE <UNPACKPANEL>
- EIGEN
- EIGENVALUE
produces a scree plot of eigenvalues and a variance-explained plot. By default, both plots are produced in a panel. Specify PLOTS= SCREE(UNPACKPANEL) to produce each plot in a separate panel. This plot is produced by default unless the CV= option is specified.
- PREFIX=name
specifies a prefix for naming the principal component scores in the OUT= data set. By default, the names are Prin1, Prin2, ..., Prin
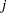 . If you specify PREFIX=ABC, the components are named ABC1, ABC2, ABC3, and so on. The number of characters in the prefix plus the number of digits in
. If you specify PREFIX=ABC, the components are named ABC1, ABC2, ABC3, and so on. The number of characters in the prefix plus the number of digits in 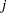 should not exceed the current name length defined by the VALIDVARNAME= system option.
should not exceed the current name length defined by the VALIDVARNAME= system option. - RPREFIX=name
specifies a prefix for naming the residual variables in the OUT= data set. By default, the prefix is R_. Residual variable names are formed by appending process variable names to the prefix. The number of characters in the prefix plus the maximum length of the process variable names should not exceed the current name length defined by the VALIDVARNAME= system option.
- STDSCORES
standardizes the principal component scores in the OUT= data set to unit variance. If you omit the STDSCORES option, the scores have variance equal to the corresponding eigenvalue. STDSCORES has no effect on the eigenvalues themselves.
- TIMEGROUP=
The TIMEGROUP= option specifies a variable in the DATA= input data set whose values indicate the time or chronological order associated with process variable measurements. This variable is not included in the principal component analysis, but you must specify this variable to compute the statistics needed to create an SPE chart when there is more than one observation per time value.
Note: This procedure is experimental.