Nonlinear Optimization Examples
Example 15.5 Profile-Likelihood-Based Confidence Intervals
This example calculates confidence intervals based on the profile likelihood for the parameters estimated in the previous example. The following introduction on profile-likelihood methods is based on the paper of Venzon and Moolgavkar (1988).
Let 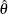 be the maximum likelihood estimate (MLE) of a parameter vector
be the maximum likelihood estimate (MLE) of a parameter vector 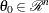 and let
and let 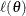 be the log-likelihood function defined for parameter values
be the log-likelihood function defined for parameter values 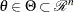 .
.
The profile-likelihood method reduces 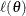 to a function of a single parameter of interest,
to a function of a single parameter of interest, 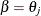 , where
, where 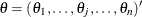 , by treating the others as nuisance parameters and maximizing over them. The profile likelihood for
, by treating the others as nuisance parameters and maximizing over them. The profile likelihood for 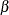 is defined as
is defined as
![\[ \tilde{\ell }_ j(\beta ) = \max _{\theta \in \Theta _ j(\beta )} \ell (\theta ) \]](images/imlug_nonlinearoptexpls0323.png)
where 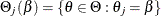 . Define the complementary parameter set
. Define the complementary parameter set 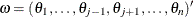 and
and 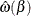 as the optimizer of
as the optimizer of 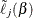 for each value of
for each value of 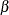 . Of course, the maximum of function
. Of course, the maximum of function 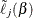 is located at
is located at 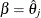 . The profile-likelihood-based confidence interval for parameter
. The profile-likelihood-based confidence interval for parameter 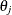 is defined as
is defined as
![\[ \{ \beta : \ell (\hat{\theta }) - \tilde{\ell }_ j(\beta ) \leq \frac{1}{2}q_1(1-\alpha ) \} \]](images/imlug_nonlinearoptexpls0330.png)
where 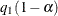 is the
is the 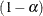 th quantile of the
th quantile of the 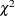 distribution with one degree of freedom. The points
distribution with one degree of freedom. The points 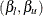 are the endpoints of the profile-likelihood-based confidence interval for parameter
are the endpoints of the profile-likelihood-based confidence interval for parameter 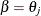 . The points
. The points 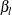 and
and 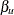 can be computed as the solutions of a system of n nonlinear equations
can be computed as the solutions of a system of n nonlinear equations 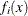 in n parameters, where
in n parameters, where 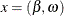 :
:
![\[ \left[ \begin{array}{c} \ell (\theta ) - \ell ^* \\ \frac{\partial \ell }{\partial \omega } (\theta ) \end{array} \right] = 0 \]](images/imlug_nonlinearoptexpls0339.png)
where 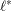 is the constant threshold
is the constant threshold 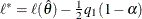 . The first of these n equations defines the locations
. The first of these n equations defines the locations 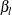 and
and 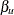 where the function
where the function 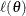 cuts
cuts 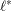 , and the remaining
, and the remaining 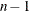 equations define the optimality of the
equations define the optimality of the 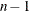 parameters in
parameters in 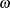 . Jointly, the n equations define the locations
. Jointly, the n equations define the locations 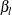 and
and 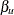 where the function
where the function 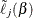 cuts the constant threshold
cuts the constant threshold 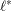 , which is given by the roots of
, which is given by the roots of 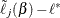 . Assuming that the two solutions
. Assuming that the two solutions 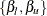 exist (they do not if the quantile
exist (they do not if the quantile 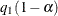 is too large), this system of n nonlinear equations can be solved by minimizing the sum of squares of the n functions
is too large), this system of n nonlinear equations can be solved by minimizing the sum of squares of the n functions 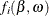 :
:
![\[ F = \frac{1}{2} \sum _{i=1}^ n f_ i^2(\beta ,\omega ) \]](images/imlug_nonlinearoptexpls0347.png)
For a solution of the system of n nonlinear equations to exist, the minimum value of the convex function F must be zero.
The following statements defines the module for the system of 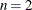 nonlinear equations to be solved in terms of the modules that are defined in the previous section:
nonlinear equations to be solved in terms of the modules that are defined in the previous section:
start f_plwei2(x) global(carcin,ipar,lstar); /* use x[1]=sig, x[2]=c, thet */ like = f_weib2(x); grad = g_weib2(x); grad[ipar] = like - lstar; return(grad`); finish f_plwei2;
The following statements implements the Levenberg-Marquardt algorithm with the NLPLM subroutine to solve the system of two
equations for the left and right endpoints of the interval. The starting point is the optimizer 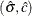 , as computed in the previous example, moved toward the left or right endpoint of the interval by an initial step (refer to
Venzon and Moolgavkar (1988)). This forces the algorithm to approach the specified endpoint.
The results, shown in Output 15.5.1, are close to the results shown in Output 15.4.2.
, as computed in the previous example, moved toward the left or right endpoint of the interval by an initial step (refer to
Venzon and Moolgavkar (1988)). This forces the algorithm to approach the specified endpoint.
The results, shown in Output 15.5.1, are close to the results shown in Output 15.4.2.
/* quantile of chi**2 distribution */
chqua = cinv(1-prob,1); lstar = fopt - .5 * chqua;
/* set number of equations = 2, and print parameter = 0 */
optn = {2 0}; /* Dont print in NLPLM */
do ipar = 1 to 2;
/* Compute initial step: */
/* Choose (alfa,delt) to go in right direction */
/* Venzon & Moolgavkar (1988), p.89 */
if ipar=1 then ind = 2; else ind = 1;
delt = - inv(hes2[ind,ind]) * hes2[ind,ipar];
alfa = - (hes2[ipar,ipar] - delt` * hes2[ind,ipar]);
if alfa > 0 then do;
alfa = .5 * sqrt(chqua / alfa);
end; else do;
print "Bad alpha";
alfa = .1 * xopt[ipar];
end;
if ipar=1 then delt = 1 // delt;
else delt = delt // 1;
/* Get upper end of interval */
x0 = xopt + alfa * delt;
/* set lower bound to optimal value */
con2 = con; con2[1,ipar] = xopt[ipar];
CALL NLPLM(rc,xres,"f_plwei2",x0,optn,con2);
f = f_plwei2(xres); s = ssq(f);
if (s < 1.e-6) then xub[ipar] = xres[ipar];
else xub[ipar] = .;
/* Get lower end of interval */
x0 = xopt - alfa * delt;
/* reset lower bound and set upper bound to optimal value */
con2[1,ipar] = con[1,ipar]; con2[2,ipar] = xopt[ipar];
CALL NLPLM(rc,xres,"f_plwei2",x0,optn,con2);
f = f_plwei2(xres); s = ssq(f);
if (s < 1.e-6) then xlb[ipar] = xres[ipar];
else xlb[ipar] = .;
end;
results = xlb || xopt || xub;
print results[L="Profile-Likelihood Confidence Interval"
c={"Lower Bound" "Estimate" "UpperBound"}
r={"sigma" "c"}];
Output 15.5.1: Confidence Interval Based on Profile Likelihood