The FAMILY Procedure
PROC FAMILY <options> ;
You can specify the following options in the PROC FAMILY statement.
- COMBINE
-
specifies that the combined versions of the S-TDT and SDT be performed. Thus, families containing parental genotypes can be analyzed under certain conditions by using the TDT; otherwise the specified sibling test is performed. Note that if TDT is also being performed, the TDT is done independently of any other tests. By default, the combined versions are not used.
-
CONTCORR
CC -
specifies that a continuity correction of 0.5 should be used for the TDT, S-TDT, and RC-TDT tests in their asymptotic normal approximations. By default, no correction is used.
- DATA=SAS-data-set
-
names the input SAS data set to be used by PROC FAMILY. The default is to use the most recently created data set.
- DELIMITER=’string’
-
indicates the string that is used to separate the two alleles that compose the genotypes contained in the variables specified in the VAR statement. This option is ignored if GENOCOL is not specified.
- GENOCOL
-
indicates that columns specified in the VAR statement contain genotypes instead of alleles. When this option is specified, there is one column per marker. The genotypes must consist of the two alleles separated by a delimiter.
-
MULT=JOINT
MULT=MAX -
specifies which multiallelic version of the TDT, S-TDT, SDT, and RC-TDT tests should be performed. The joint version of the multiallelic tests combines the analyses for each allele at a marker into one overall test statistic, with degrees of freedom (df) corresponding to the number of alleles at the marker. The max version of the multiallelic tests determines whether there is at least one allele with a significant test statistic, using the maximum 1 df statistic over all alleles with a multiple testing adjustment made. By default, the joint version of the multiallelic tests is performed. This option has no effect on biallelic markers.
- NDATA=SAS-data-set
-
names the input SAS data set containing names, or identifiers, for the markers used in the output. There must be a
NAMEvariable in this data set, which should contain the same number of rows as there are markers in the input data set specified in the DATA= option. When there are fewer rows than there are markers, markers without a name are named using the PREFIX= option. Likewise, if there is no NDATA= data set specified, the PREFIX= option is used. If both the VAR and XLVAR statements are specified, names are first used for the markers in the VAR statement, then for the X-linked markers. - OUTQ=SAS-data-set
-
names the output SAS data set containing all the variables from the input data set in addition to the allelic transmission scores at each marker allele to be used in testing for association and linkage with a quantitative trait. When this option is used, the TRAIT statement is not required.
- OUTSTAT=SAS-data-set
-
names the output SAS data set containing the
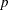 -values for the tests specified in the PROC FAMILY statement. When this option is omitted, an output data set is created by
default and named according to the DATAn convention.
-values for the tests specified in the PROC FAMILY statement. When this option is omitted, an output data set is created by
default and named according to the DATAn convention.
- PERMS=number
-
indicates that Monte Carlo estimates of exact
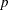 -values for the family-based tests should be calculated using permutation samples instead of the
-values for the family-based tests should be calculated using permutation samples instead of the 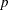 -values from the asymptotic
-values from the asymptotic 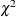 distribution. Large values of number (10,000 or more) are usually recommended for accuracy, but long execution times can result, particularly with large data
sets. When this option is omitted, no permutations are performed and
distribution. Large values of number (10,000 or more) are usually recommended for accuracy, but long execution times can result, particularly with large data
sets. When this option is omitted, no permutations are performed and 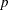 -values from the asymptotic
-values from the asymptotic 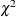 distribution are reported.
distribution are reported.
- PREFIX=prefix
-
specifies a prefix to use in constructing names for marker variables in all output. For example, if PREFIX=VAR, the names of the variables are VAR1, VAR2, …, VARn. Note that this option is ignored when the NDATA= option is specified, unless there are fewer names in the NDATA data set than there are markers. If this option is omitted, PREFIX=M is the default when variables contain alleles; if GENOCOL is specified, then the names of the variables specified in the VAR statement are used as the marker names.
- RCTDT
-
requests that the reconstruction-combined TDT (RC-TDT) be performed. If none of the four test options (RCTDT, SDT, STDT, or TDT) are specified, then all four tests are performed by default. Note that error checking is always performed on families with at least one untyped parent in order to determine whether or not reconstruction of parental genotypes can be attempted.
- SDT
-
requests that the SDT, a nonparametric alternative to the S-TDT, be performed. If none of the four test options (RCTDT, SDT, STDT, or TDT) are specified, then all four tests are performed by default. The COMBINE option can be used with this test to indicate that the combined version of the SDT should be performed.
- SEED=number
-
specifies the initial seed for the random number generator used for permuting the data to calculate estimates of exact
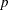 -values. This option is ignored if PERMS= is not specified. The value for number must be an integer; the computer clock time is used if the option is omitted or an integer less than or equal to 0 is specified.
For more details about seed values, see SAS Language Reference: Concepts.
-values. This option is ignored if PERMS= is not specified. The value for number must be an integer; the computer clock time is used if the option is omitted or an integer less than or equal to 0 is specified.
For more details about seed values, see SAS Language Reference: Concepts.
- SHOWALL
-
indicates that all families and markers should be included in the “Family Summary” table. When this option is omitted, a family is included in the table only for a marker where there is a genotype error according to a Mendelian inconsistency.
- STDT
-
requests that the sibling TDT (S-TDT), which analyzes data from sibships, be performed. If none of the four test options (RCTDT, SDT, STDT, or TDT) are specified, then all four tests are performed by default. The COMBINE option can be used with this test to indicate that the combined version of the S-TDT should be performed.
- TDT
-
requests that the original TDT be performed. If none of the four test options (RCTDT, SDT, STDT, or TDT) are specified, then all four tests are performed by default.
