| The HAPLOTYPE Procedure |
Example 8.5 Creating a Data Set for a Regression Model
Another approach to testing haplotypes for association with a phenotype uses a regression model, which can be more powerful than the omnibus chi-square test performed in PROC HAPLOTYPE (Schaid et al. 2002; Zaykin et al. 2002). The output data set produced by PROC HAPLOTYPE can easily be transformed into one that can be used by one of the regression procedures offered by SAS/STAT. This approach can be used for quantitative traits as well as binary or ordinal traits.
Here is an example data set that can be analyzed using PROC HAPLOTYPE:
data alleles;
input (a1-a6) ($) disease;
datalines;
A a B B c C 1
A A B b c C 1
a A B b c c 0
A A B B c C 1
A A b B c C 1
A A B b C c 0
A a b B C c 1
A A b B C c 1
A a B B c c 1
a a B b c c 0
A A B B C C 1
A A B B c c 1
a A b b c c 0
A A B B c c 1
A A b b c c 0
A A b B c C 0
A A B b c C 1
A a b B c c 1
A a B B c C 1
A A b b C C 0
A A B B C C 1
A A b B C c 1
A A b B c C 1
a A B b C c 0
A a B B C C 0
A A B B C c 1
A A B b C c 0
A A B B c C 1
a A B b C C 1
A a B b C c 1
A A B b c C 1
A a B B c c 1
A A B b C c 1
a A B b C c 1
A A B b C C 1
A a B B C C 1
a A B b C c 0
a A b B C C 0
A A B b c C 1
a A B b c c 0
A A B B C C 0
A A B B c c 1
A a B B C c 1
;
The following code creates an output data set containing individuals’ probabilities of having particular haplotype pairs, with the ID statement and OUTID option indicating that this data set includes the disease variable from the input data set and a unique identifier for each individual assigned by PROC HAPLOTYPE, respectively. An omnibus test for association between the three markers and disease status is also performed.
proc haplotype data=alleles out=out outid;
var a1-a6;
trait disease;
id disease;
run;
This code executes the omnibus marker-trait association test whose  -value is given by the chi-square distribution.
-value is given by the chi-square distribution.
| Test for Marker-Trait Association | ||||||
|---|---|---|---|---|---|---|
| Trait Number |
Trait Value |
Num Obs |
DF | LogLike | Chi-Square | Pr > ChiSq |
| 1 | 1 | 29 | 7 | -68.11558 | ||
| 2 | 0 | 14 | 7 | -37.28544 | ||
| Combined | 43 | 7 | -115.48338 | 20.1647 | 0.0052 | |
Output 8.5.1 shows that there is a significant overall association between the markers and the trait, disease status. However, the more powerful score test for regression can be implemented by using the following code to perform a test for additive effects of the marker haplotypes.
data out1;
set out;
haplotype=tranwrd(haplotype1,'-','_');
run;
data out2;
set out;
haplotype=tranwrd(haplotype2,'-','_');
run;
data outnew;
set out1 out2;
run;
proc sort data=outnew;
by haplotype;
run;
data outnew2;
set outnew;
lagh=lag(haplotype);
if haplotype ne lagh then num+1;
hapname=compress("H"||num,' ');
run;
proc sort data=outnew2;
by _id_ haplotype;
run;
data outt;
set outnew2;
by _id_ haplotype;
if first.haplotype then totprob=prob/2;
else totprob+prob/2;
if last.haplotype;
run;
proc transpose data=outt out=outreg(drop=_NAME_) ;
id hapname;
idlabel haplotype;
var totprob;
by _id_ disease;
run;
data htr;
set outreg;
array h{8};
do i=1 to 8;
if h{i}=. then h{i}=0;
end;
keep _id_ disease h1-h8;
run;
proc print data=htr noobs round label;
run;
proc logistic data=htr descending;
model disease = h1-h8 / selection=stepwise;
run;
This SAS code produces a data set htr from the output data set of PROC HAPLOTYPE that contains the variables needed to be able to perform a regression analysis. There is now one column for each possible haplotype in the sample, with each column containing the haplotype’s frequency, or probability, within an individual.
The data set shown in Output 8.5.2 can now be used in one of the regression procedures offered by SAS/STAT. In this example, since the trait is binary, the LOGISTIC procedure can be used to perform a regression on the variable disease. The REG procedure could be used in a similar manner to analyze a quantitative trait.
| The HAPLOTYPE Procedure |
| Tests for Haplotype-Trait Association |
| Individual ID | disease | A_B_C | A_B_c | a_B_C | a_B_c | A_b_C | A_b_c | a_b_c | a_b_C |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0.29 | 0.21 | 0.21 | 0.29 | 0.00 | 0.00 | 0.00 | 0 |
| 2 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 3 | 0 | 0.00 | 0.27 | 0.00 | 0.23 | 0.00 | 0.23 | 0.27 | 0 |
| 4 | 1 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 5 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 6 | 0 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 7 | 1 | 0.22 | 0.00 | 0.13 | 0.15 | 0.15 | 0.13 | 0.22 | 0 |
| 8 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 9 | 1 | 0.00 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0 |
| 10 | 0 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.50 | 0 |
| 11 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 12 | 1 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 13 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 | 0.50 | 0 |
| 14 | 1 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 15 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0 |
| 16 | 0 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 17 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 18 | 1 | 0.00 | 0.27 | 0.00 | 0.23 | 0.00 | 0.23 | 0.27 | 0 |
| 19 | 1 | 0.29 | 0.21 | 0.21 | 0.29 | 0.00 | 0.00 | 0.00 | 0 |
| 20 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0 |
| 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 22 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 23 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 24 | 0 | 0.22 | 0.00 | 0.13 | 0.15 | 0.15 | 0.13 | 0.22 | 0 |
| 25 | 0 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 26 | 1 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 27 | 0 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 28 | 1 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 29 | 1 | 0.01 | 0.00 | 0.49 | 0.00 | 0.49 | 0.00 | 0.00 | 0 |
| 30 | 1 | 0.22 | 0.00 | 0.13 | 0.15 | 0.15 | 0.13 | 0.22 | 0 |
| 31 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 32 | 1 | 0.00 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0 |
| 33 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 34 | 1 | 0.22 | 0.00 | 0.13 | 0.15 | 0.15 | 0.13 | 0.22 | 0 |
| 35 | 1 | 0.50 | 0.00 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0 |
| 36 | 1 | 0.50 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 37 | 0 | 0.22 | 0.00 | 0.13 | 0.15 | 0.15 | 0.13 | 0.22 | 0 |
| 38 | 0 | 0.01 | 0.00 | 0.49 | 0.00 | 0.49 | 0.00 | 0.00 | 0 |
| 39 | 1 | 0.27 | 0.23 | 0.00 | 0.00 | 0.23 | 0.27 | 0.00 | 0 |
| 40 | 0 | 0.00 | 0.27 | 0.00 | 0.23 | 0.00 | 0.23 | 0.27 | 0 |
| 41 | 0 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 42 | 1 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 |
| 43 | 1 | 0.29 | 0.21 | 0.21 | 0.29 | 0.00 | 0.00 | 0.00 | 0 |
Output 8.5.3 shows two of the tables produced by PROC LOGISTIC. The first one displays the test of the global null hypothesis, 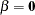 . You can see that the score test indicates a significant association between the haplotypes at the three markers and disease status. In particular, the second table shows that as a result of the stepwise selection, the haplotype H8 (a-b-c) has a statistically significant effect on disease status. This is an example of how a regression analysis can be used to detect association in a similar manner to the LRT implemented by PROC HAPLOTYPE.
. You can see that the score test indicates a significant association between the haplotypes at the three markers and disease status. In particular, the second table shows that as a result of the stepwise selection, the haplotype H8 (a-b-c) has a statistically significant effect on disease status. This is an example of how a regression analysis can be used to detect association in a similar manner to the LRT implemented by PROC HAPLOTYPE.
| Testing Global Null Hypothesis: BETA=0 | |||
|---|---|---|---|
| Test | Chi-Square | DF | Pr > ChiSq |
| Likelihood Ratio | 6.1962 | 1 | 0.0128 |
| Score | 6.3995 | 1 | 0.0114 |
| Wald | 4.9675 | 1 | 0.0258 |
| Analysis of Maximum Likelihood Estimates | |||||
|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error |
Wald Chi-Square |
Pr > ChiSq |
| Intercept | 1 | 1.1986 | 0.4058 | 8.7224 | 0.0031 |
| H8 | 1 | -6.3249 | 2.8378 | 4.9675 | 0.0258 |
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.