| The GENESELECT Procedure |
| IMPORTANCE Statement |
- IMPORTANCE </ options> ;
The IMPORTANCE statement implements an observation-based approach to evaluate the importance of a variable or of a pair of variables to the predictions of the model.
For each observation, the value of the variable or pair of variables being evaluated is rendered uninformative. The IMPORTANCE statement outputs the prediction once using the actual value and a second time using the uninformative value. The difference between the two predictions shows the dependence of the prediction on the variable or pair of variables being evaluated. The differences for all the observations can be plotted against the actual variable value or observation number to explore where the dependence is stronger or weaker.
The observation-based importance differs from the split-based importance computed in the IMPORTANCE= option of the SAVE statement. The latter importance is based on the contribution a variable makes in reducing the residual sum of squares.
The IMPORTANCE statement can be repeated.
- DATA=SAS-data-set
names the input data set. If the DATA= option is absent, the procedure uses the training data.
- N2WAY=m n
requests to evaluate the best
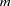 variables paired with the best
variables paired with the best  variables, where the term "best" here refers to the split-based variable importance rankings computed in the IMPORTANCE= option of the SAVE statement. If
variables, where the term "best" here refers to the split-based variable importance rankings computed in the IMPORTANCE= option of the SAVE statement. If  is missing, then
is missing, then  is set to
is set to 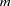 . The default value of
. The default value of 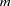 and
and  is 0. When the procedure evaluates a pair of variables, it also evaluates the two variables individually and outputs results as if the variables were specified in the VAR= option.
is 0. When the procedure evaluates a pair of variables, it also evaluates the two variables individually and outputs results as if the variables were specified in the VAR= option. - NVARS=n
requests to evaluate the best
 variables as ranked by the split-based variable importance computed in the IMPORTANCE= option of the SAVE statement. If the N2WAY=, NVARS=, and VAR= options are absent, then the procedure assumes NVARS=5.
variables as ranked by the split-based variable importance computed in the IMPORTANCE= option of the SAVE statement. If the N2WAY=, NVARS=, and VAR= options are absent, then the procedure assumes NVARS=5. - OUT=SAS-data-set
names the output data set to contain the scored data. If the OUT= option is absent, the procedure creates a data set name using the DATA
 convention.
convention. The OUT= data set in the IMPORTANCE has the same variables as the OUT= data set in the SCORE data set, plus one or two more, _INPUT1_ and _INPUT2_, that contain the name of a variable whose values were treated as uninformative when making the predictions. If _INPUT1_ is blank, then _INPUT2_ is blank and the predictions are the same as in the OUT= data set of the SCORE statement.
The OUT= data set becomes very large if many variables are being evaluated. The number of observations in the OUT= data set equals the number of variables and pairs of variables being evaluated plus one times the number of observations in the data set. Specify OUT=_NULL_ to avoid creating a scored data set.
- OUTFIT=SAS-data-set
names the output data set to contain the fit statistics. The number of observations in the OUTFIT= data set equals the number of variables and pairs of variables being evaluated plus one.
The OUTFIT= data set in the IMPORTANCE has the same variables as the OUTFIT= data set in the SCORE data set, plus one or two more, _INPUT1_ and _INPUT2_, that contain the name of a variable whose values were treated as uninformative when computating the statistics. If _INPUT1_ is blank, then _INPUT2_ is blank and the statistics are the same as in the OUTFIT= data set of the SCORE statement.
- VAR=(varlist)
specifies variables and pairs of variables to evaluate. Varlist is a list of variable names optionally containing asterisks to indicate a pair of variables. Variables on the left or right of an asterisk may be grouped within square brackets. Brackets may not be nested. Parentheses must enclose the list, varlist.
When a procedure evaluates a pair of variables, it also evaluates the two variables individually and outputs the results. For example, the following varlist would specify variables A, B, C, D, E, and pairs of variables, B-C, D-E, D-C, and E-C:
A B*C [D E] * [E C]
Note: This procedure is experimental.
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.