The SURVEYPHREG Procedure
- Overview
- Getting Started
-
Syntax
 PROC SURVEYPHREG StatementBY StatementCLASS StatementCLUSTER StatementDOMAIN StatementESTIMATE StatementFREQ StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementOUTPUT StatementProgramming StatementsREPWEIGHTS StatementSLICE StatementSTORE StatementSTRATA StatementTEST StatementWEIGHT Statement
PROC SURVEYPHREG StatementBY StatementCLASS StatementCLUSTER StatementDOMAIN StatementESTIMATE StatementFREQ StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementOUTPUT StatementProgramming StatementsREPWEIGHTS StatementSLICE StatementSTORE StatementSTRATA StatementTEST StatementWEIGHT Statement -
Details
 Notation and EstimationFailure Time DistributionTime and CLASS Variables UsagePartial Likelihood Function for the Cox ModelSpecifying the Sample DesignMissing ValuesVariance EstimationDomain AnalysisHypothesis Tests, Confidence Intervals, and ResidualsOutput Data SetsDisplayed OutputODS Table NamesODS Graphics
Notation and EstimationFailure Time DistributionTime and CLASS Variables UsagePartial Likelihood Function for the Cox ModelSpecifying the Sample DesignMissing ValuesVariance EstimationDomain AnalysisHypothesis Tests, Confidence Intervals, and ResidualsOutput Data SetsDisplayed OutputODS Table NamesODS Graphics -
Examples

- References
Jackknife Method
The jackknife method of variance estimation deletes one PSU at a time from the full sample to create replicates. This method
is also known as the delete-1 jackknife method because it deletes exactly one PSU in every replicate. The total number of
replicates R is the same as the total number of PSUs. In each replicate, the sampling weights of the remaining PSUs are modified by the
jackknife coefficient  . The modified weights are called replicate weights.
. The modified weights are called replicate weights.
Let PSU i in stratum  be omitted for the rth replicate; then the jackknife coefficient and replicate weights are computed as
be omitted for the rth replicate; then the jackknife coefficient and replicate weights are computed as
![\[ \alpha _ r = \left\{ \begin{array}{ll} \frac{ n_{h_ r} -1 }{n_{h_ r}} & \text {for a stratified design} \\[0.1in] \frac{R-1}{R} & \text {for designs without stratification} \end{array} \right. \]](images/statug_surveyphreg0105.png)
and
![\[ w_{hij}^{(r)} = \left\{ \begin{array}{ll} w_{hij} & \text {if observation unit } j \text { is not in donor stratum } h_ r \\ 0 & \text {if observation unit } j \text { is in PSU } i \text { of donor stratum } h_ r \\ w_{hij} / \alpha _ r & \text {if observation unit } j \text { is not in PSU } i \text { but in donor stratum } h_ r \end{array} \right. \]](images/statug_surveyphreg0106.png)
You can use the VARMETHOD=JACKKNIFE(OUTJKCOEFS=) method-option to store the jackknife coefficients in a SAS data set and use the VARMETHOD=JACKKNIFE(OUTWEIGHTS=) method-option to store the replicate weights in a SAS data set.
If you provide your own replicate weights with a REPWEIGHTS statement, then you can also provide corresponding jackknife coefficients
with the JKCOEFS=
option. If you provide replicate weights with a REPWEIGHTS statement but do not provide jackknife coefficients, then the
procedure uses 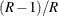 as the default jackknife coefficient for every replicate, where R is the total number of replicates.
as the default jackknife coefficient for every replicate, where R is the total number of replicates.
Let 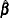 be the estimated proportional hazards regression coefficients from the full sample, and let
be the estimated proportional hazards regression coefficients from the full sample, and let  be the estimated regression coefficients for the rth replicate. PROC SURVEYPHREG estimates the covariance matrix of
be the estimated regression coefficients for the rth replicate. PROC SURVEYPHREG estimates the covariance matrix of 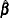 by
by
![\[ \widehat{\mb{V}}(\hat{\bbeta }) = \sum _{r=1}^ R \alpha _ r \left( {\hat{\bbeta }_ r} - \hat{\bbeta } \right) \left( {\hat{\bbeta }_ r} - \hat{\bbeta } \right)’ \]](images/statug_surveyphreg0107.png)
with  degrees of freedom, where R is the number of replicates and H is the number of strata, or R – 1 when there is no stratification.
degrees of freedom, where R is the number of replicates and H is the number of strata, or R – 1 when there is no stratification.
If you specify the CENTER=REPLICATES
method-option, then PROC SURVEYPHREG computes the covariance matrix of 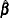 by
by
![\[ \widehat{\mb{V}}(\hat{\bbeta }) = \sum _{r=1}^ R \alpha _ r \left( {\hat{\bbeta }_ r} - \overline{\hat{\bbeta }_ r} \right) \left( {\hat{\bbeta }_ r} - \overline{\hat{\bbeta }_ r} \right)’ \]](images/statug_surveyphreg0109.png)
where  is the average of the replicate estimates as follows:
is the average of the replicate estimates as follows:
![\[ \overline{\hat{\bbeta }_ r} = \frac{1}{R} \sum _{r=1}^ R \hat{\bbeta _ r} \]](images/statug_surveyphreg0091.png)
If one or more components of  cannot be calculated for some replicates, then the variance estimator uses only the replicates for which the proportional
hazards regression coefficients can be estimated. Estimability and nonconvergence are two common reasons why
cannot be calculated for some replicates, then the variance estimator uses only the replicates for which the proportional
hazards regression coefficients can be estimated. Estimability and nonconvergence are two common reasons why  might not be available for a replicate sample even if
might not be available for a replicate sample even if 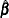 is defined for the full sample. Let
is defined for the full sample. Let 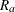 be the number of replicates where
be the number of replicates where  are available, and let
are available, and let 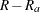 be the number of replicates where
be the number of replicates where  are not available. Without loss of generality, assume that
are not available. Without loss of generality, assume that  is available only for the first
is available only for the first 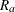 replicates; then the jackknife variance estimator is
replicates; then the jackknife variance estimator is
![\[ \widehat{\mb{V}}(\hat{\bbeta }) = \sum _{r=1}^{R_ a} \alpha _ r \left( {\hat{\bbeta }_ r} - \hat{\bbeta } \right) \left( {\hat{\bbeta }_ r} - \hat{\bbeta } \right)’ \]](images/statug_surveyphreg0110.png)
with  degrees of freedom, where H is the number of strata. Alternatively, you can use the VADJUST=AVGREPSS
option in the MODEL statement to use the average sum of squares for the invalid replicate samples. See Variance Adjustment Factors for details.
degrees of freedom, where H is the number of strata. Alternatively, you can use the VADJUST=AVGREPSS
option in the MODEL statement to use the average sum of squares for the invalid replicate samples. See Variance Adjustment Factors for details.