The GLIMMIX Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GLIMMIX StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementCOVTEST StatementEFFECT StatementESTIMATE StatementFREQ StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementOUTPUT StatementPARMS StatementRANDOM StatementSLICE StatementSTORE StatementWEIGHT StatementProgramming StatementsUser-Defined Link or Variance Function
PROC GLIMMIX StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementCOVTEST StatementEFFECT StatementESTIMATE StatementFREQ StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementOUTPUT StatementPARMS StatementRANDOM StatementSLICE StatementSTORE StatementWEIGHT StatementProgramming StatementsUser-Defined Link or Variance Function -
Details
 Generalized Linear Models TheoryGeneralized Linear Mixed Models TheoryGLM Mode or GLMM ModeStatistical Inference for Covariance ParametersDegrees of Freedom MethodsEmpirical Covariance ("Sandwich") EstimatorsExploring and Comparing Covariance MatricesProcessing by SubjectsRadial Smoothing Based on Mixed ModelsOdds and Odds Ratio EstimationParameterization of Generalized Linear Mixed ModelsResponse-Level Ordering and ReferencingComparing the GLIMMIX and MIXED ProceduresSingly or Doubly Iterative FittingDefault Estimation TechniquesDefault OutputNotes on Output StatisticsODS Table NamesODS Graphics
Generalized Linear Models TheoryGeneralized Linear Mixed Models TheoryGLM Mode or GLMM ModeStatistical Inference for Covariance ParametersDegrees of Freedom MethodsEmpirical Covariance ("Sandwich") EstimatorsExploring and Comparing Covariance MatricesProcessing by SubjectsRadial Smoothing Based on Mixed ModelsOdds and Odds Ratio EstimationParameterization of Generalized Linear Mixed ModelsResponse-Level Ordering and ReferencingComparing the GLIMMIX and MIXED ProceduresSingly or Doubly Iterative FittingDefault Estimation TechniquesDefault OutputNotes on Output StatisticsODS Table NamesODS Graphics -
Examples
 Binomial Counts in Randomized BlocksMating Experiment with Crossed Random EffectsSmoothing Disease Rates; Standardized Mortality RatiosQuasi-likelihood Estimation for Proportions with Unknown DistributionJoint Modeling of Binary and Count DataRadial Smoothing of Repeated Measures DataIsotonic Contrasts for Ordered AlternativesAdjusted Covariance Matrices of Fixed EffectsTesting Equality of Covariance and Correlation MatricesMultiple Trends Correspond to Multiple Extrema in Profile LikelihoodsMaximum Likelihood in Proportional Odds Model with Random EffectsFitting a Marginal (GEE-Type) ModelResponse Surface Comparisons with Multiplicity AdjustmentsGeneralized Poisson Mixed Model for Overdispersed Count DataComparing Multiple B-SplinesDiallel Experiment with Multimember Random EffectsLinear Inference Based on Summary DataWeighted Multilevel Model for Survey DataQuadrature Method for Multilevel Model
Binomial Counts in Randomized BlocksMating Experiment with Crossed Random EffectsSmoothing Disease Rates; Standardized Mortality RatiosQuasi-likelihood Estimation for Proportions with Unknown DistributionJoint Modeling of Binary and Count DataRadial Smoothing of Repeated Measures DataIsotonic Contrasts for Ordered AlternativesAdjusted Covariance Matrices of Fixed EffectsTesting Equality of Covariance and Correlation MatricesMultiple Trends Correspond to Multiple Extrema in Profile LikelihoodsMaximum Likelihood in Proportional Odds Model with Random EffectsFitting a Marginal (GEE-Type) ModelResponse Surface Comparisons with Multiplicity AdjustmentsGeneralized Poisson Mixed Model for Overdispersed Count DataComparing Multiple B-SplinesDiallel Experiment with Multimember Random EffectsLinear Inference Based on Summary DataWeighted Multilevel Model for Survey DataQuadrature Method for Multilevel Model - References
Residual-Based Estimators
The GLIMMIX procedure can compute the classical sandwich estimator of the covariance matrix of the fixed effects, as well as several bias-adjusted estimators. This requires that the model is either an (overdispersed) GLM or a GLMM that can be processed by subjects (see the section Processing by Subjects).
Consider a statistical model of the form
![\[ \mb{Y} = \bmu + \bepsilon , \qquad \bepsilon \sim (\mb{0},\bSigma ) \]](images/statug_glimmix0793.png)
The general expression of a sandwich covariance estimator is then
![\[ c \times \widehat{\bOmega } \left( \sum _{i=1}^ m \mb{A}_ i \widehat{\mb{D}}_ i’ \widehat{\bSigma }_ i^{-1} \mb{F}_ i’ \mb{e}_ i\mb{e}_ i’ \mb{F}_ i \widehat{\bSigma }_ i^{-1} \widehat{\mb{D}}_ i \mb{A}_ i \right) \widehat{\bOmega } \]](images/statug_glimmix0794.png)
where 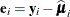 ,
, 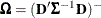 .
.
For a GLMM estimated by one of the pseudo-likelihood techniques that involve linearization, you can make the following substitutions:
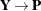 ,
, 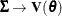 ,
, 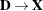 ,
, 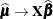 . These matrices are defined in the section Pseudo-likelihood Estimation Based on Linearization.
. These matrices are defined in the section Pseudo-likelihood Estimation Based on Linearization.
The various estimators computed by the GLIMMIX procedure differ in the choice of the constant c and the matrices 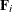 and
and 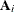 . You obtain the classical estimator, for example, with c = 1, and
. You obtain the classical estimator, for example, with c = 1, and 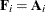 equal to the identity matrix.
equal to the identity matrix.
The EMPIRICAL= ROOT estimator of Kauermann and Carroll (2001) is based on the approximation
![\[ \mr{Var}\left[ \mb{e}_ i\mb{e}_ i’ \right] \approx (\mb{I} - \mb{H}_ i)\bSigma _ i \]](images/statug_glimmix0802.png)
where 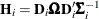 . The EMPIRICAL=
FIRORES estimator is based on the approximation
. The EMPIRICAL=
FIRORES estimator is based on the approximation
![\[ \mr{Var}\left[ \mb{e}_ i\mb{e}_ i’ \right] \approx (\mb{I} - \mb{H}_ i)\bSigma _ i(\mb{I} - \mb{H}_ i’) \]](images/statug_glimmix0804.png)
of Mancl and DeRouen (2001). Finally, the EMPIRICAL=
FIROEEQ estimator is based on approximating an unbiased estimating equation (Fay and Graubard 2001). For this estimator, 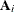 is a diagonal matrix with entries
is a diagonal matrix with entries
![\[ [\mb{A}_ i]_{jj} = \left(1 - \mr{min}\{ r,[\mb{Q}]_{jj} \} \right)^{-1/2} \]](images/statug_glimmix0805.png)
where 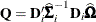 . The optional number
. The optional number 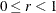 is chosen to provide an upper bound on the correction factor. For r = 0, the classical sandwich estimator results. PROC GLIMMIX chooses as default value
is chosen to provide an upper bound on the correction factor. For r = 0, the classical sandwich estimator results. PROC GLIMMIX chooses as default value 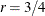 . The diagonal entries of
. The diagonal entries of 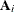 are then no greater than 2.
are then no greater than 2.
Table 45.24 summarizes the components of the computation for the GLMM based on linearization, where m denotes the number of subjects and k is the rank of 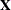 .
.
Table 45.24: Empirical Covariance Estimators for a Linearized GLMM
|
EMPIRICAL= |
c |
|
|
|---|---|---|---|
|
CLASSICAL |
1 |
|
|
|
DF |
|
|
|
|
ROOT |
1 |
|
|
|
FIRORES |
1 |
|
|
|
FIROEEQ(r) |
1 |
|
|
Computation of an empirical variance estimator requires that the data can be processed by independent sampling units. This is always the case in GLMs. In this case, m equals the sum of all frequencies. In GLMMs, the empirical estimators require that the data consist of multiple subjects. In that case, m equals the number of subjects as per the "Dimensions" table. The following section discusses how the GLIMMIX procedure determines whether the data can be processed by subjects. The section GLM Mode or GLMM Mode explains how PROC GLIMMIX determines whether a model is fit in GLM mode or in GLMM mode.
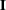
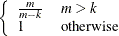
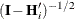
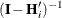
![$\mr{Diag}\{ ( 1 - \mr{min}\{ r,[\mb{Q}]_{jj} \} )^{-1/2} \} $](images/statug_glimmix0812.png)