The GLIMMIX Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GLIMMIX StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementCOVTEST StatementEFFECT StatementESTIMATE StatementFREQ StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementOUTPUT StatementPARMS StatementRANDOM StatementSLICE StatementSTORE StatementWEIGHT StatementProgramming StatementsUser-Defined Link or Variance Function
PROC GLIMMIX StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementCOVTEST StatementEFFECT StatementESTIMATE StatementFREQ StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementOUTPUT StatementPARMS StatementRANDOM StatementSLICE StatementSTORE StatementWEIGHT StatementProgramming StatementsUser-Defined Link or Variance Function -
Details
 Generalized Linear Models TheoryGeneralized Linear Mixed Models TheoryGLM Mode or GLMM ModeStatistical Inference for Covariance ParametersDegrees of Freedom MethodsEmpirical Covariance ("Sandwich") EstimatorsExploring and Comparing Covariance MatricesProcessing by SubjectsRadial Smoothing Based on Mixed ModelsOdds and Odds Ratio EstimationParameterization of Generalized Linear Mixed ModelsResponse-Level Ordering and ReferencingComparing the GLIMMIX and MIXED ProceduresSingly or Doubly Iterative FittingDefault Estimation TechniquesDefault OutputNotes on Output StatisticsODS Table NamesODS Graphics
Generalized Linear Models TheoryGeneralized Linear Mixed Models TheoryGLM Mode or GLMM ModeStatistical Inference for Covariance ParametersDegrees of Freedom MethodsEmpirical Covariance ("Sandwich") EstimatorsExploring and Comparing Covariance MatricesProcessing by SubjectsRadial Smoothing Based on Mixed ModelsOdds and Odds Ratio EstimationParameterization of Generalized Linear Mixed ModelsResponse-Level Ordering and ReferencingComparing the GLIMMIX and MIXED ProceduresSingly or Doubly Iterative FittingDefault Estimation TechniquesDefault OutputNotes on Output StatisticsODS Table NamesODS Graphics -
Examples
 Binomial Counts in Randomized BlocksMating Experiment with Crossed Random EffectsSmoothing Disease Rates; Standardized Mortality RatiosQuasi-likelihood Estimation for Proportions with Unknown DistributionJoint Modeling of Binary and Count DataRadial Smoothing of Repeated Measures DataIsotonic Contrasts for Ordered AlternativesAdjusted Covariance Matrices of Fixed EffectsTesting Equality of Covariance and Correlation MatricesMultiple Trends Correspond to Multiple Extrema in Profile LikelihoodsMaximum Likelihood in Proportional Odds Model with Random EffectsFitting a Marginal (GEE-Type) ModelResponse Surface Comparisons with Multiplicity AdjustmentsGeneralized Poisson Mixed Model for Overdispersed Count DataComparing Multiple B-SplinesDiallel Experiment with Multimember Random EffectsLinear Inference Based on Summary DataWeighted Multilevel Model for Survey DataQuadrature Method for Multilevel Model
Binomial Counts in Randomized BlocksMating Experiment with Crossed Random EffectsSmoothing Disease Rates; Standardized Mortality RatiosQuasi-likelihood Estimation for Proportions with Unknown DistributionJoint Modeling of Binary and Count DataRadial Smoothing of Repeated Measures DataIsotonic Contrasts for Ordered AlternativesAdjusted Covariance Matrices of Fixed EffectsTesting Equality of Covariance and Correlation MatricesMultiple Trends Correspond to Multiple Extrema in Profile LikelihoodsMaximum Likelihood in Proportional Odds Model with Random EffectsFitting a Marginal (GEE-Type) ModelResponse Surface Comparisons with Multiplicity AdjustmentsGeneralized Poisson Mixed Model for Overdispersed Count DataComparing Multiple B-SplinesDiallel Experiment with Multimember Random EffectsLinear Inference Based on Summary DataWeighted Multilevel Model for Survey DataQuadrature Method for Multilevel Model - References
Pseudo-likelihood Estimation for Weighted Multilevel Models
Multilevel models provide a flexible and powerful tool for the analysis of data that are observed in nested units at multiple levels. A multilevel model is a special case of generalized linear mixed models that can be handled by the GLIMMIX procedure. In proc GLIMMIX, the SUBJECT= option in the RANDOM statement identifies the clustering structure for the random effects. When the subjects of the multiple RANDOM statements are nested, the model is a multilevel model and each RANDOM statement corresponds to one level.
Using a pseudo-maximum-likelihood approach, you can extend the multilevel model framework to accommodate weights at different levels. Such an approach is very useful in analyzing survey data that arise from multistage sampling. In these sampling designs, survey weights are often constructed to account for unequal sampling probabilities, nonresponse adjustments, and poststratification.
The following survey example from a three-stage sampling design illustrates the use of multiple levels of weighting in a multilevel
model. Extending this example to models that have more than three levels is straightforward. Let 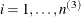 ,
, 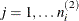 , and
, and 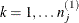 denote the indices of units at level 3, level 2, and level 1, respectively. Let superscript
denote the indices of units at level 3, level 2, and level 1, respectively. Let superscript 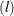 denote the lth level and
denote the lth level and 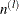 denote the number of level-l units in the sample. Assume that the first-stage cluster (level-3 unit) i is selected with probability
denote the number of level-l units in the sample. Assume that the first-stage cluster (level-3 unit) i is selected with probability 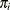 ; the second-stage cluster (level-2 unit) j is selected with probability
; the second-stage cluster (level-2 unit) j is selected with probability 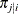 , given that the first-stage cluster i is already selected in the sample; and the third-stage unit (level-1 unit) k is selected with probability
, given that the first-stage cluster i is already selected in the sample; and the third-stage unit (level-1 unit) k is selected with probability 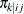 , given that the second-stage cluster j within the first-stage cluster i is already selected in the sample.
, given that the second-stage cluster j within the first-stage cluster i is already selected in the sample.
If you use the inverse selection probability weights 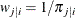 ,
, 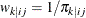 , the conditional log likelihood contribution of the first-stage cluster i is
, the conditional log likelihood contribution of the first-stage cluster i is
![\[ \log {(p(y_{i}|\gamma _{i}^{(2)},\gamma _{i}^{(3)}))}=\sum _{j=1}^{n_ i^{(2)}} {w_{j|i}} \sum _{k=1}^{n_ j^{(1)}} {w_{k|ij}} \log {( p(y_{ijk} |\gamma _{ij}^{(2)}\gamma _{i}^{(3)} ))} \]](images/statug_glimmix0697.png)
where 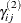 is the random-effects vector for the jth second-stage cluster,
is the random-effects vector for the jth second-stage cluster, 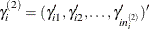 , and
, and 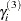 is the random-effects vector for the ith first-stage cluster.
is the random-effects vector for the ith first-stage cluster.
As with unweighted multilevel models, the adaptive quadrature method is used to compute the pseudo-likelihood of the first-stage cluster i:
![\[ p(y_{i}) = \int p(y_{i}|\gamma _{i}^{(2)},\gamma _{i}^{(3)}) p( \gamma _{i}^{(2)})p( \gamma _{i}^{(3)})d(\gamma _{i}^{(2)}) d(\gamma _{i}^{(3)}) \]](images/statug_glimmix0701.png)
The total log pseudo-likelihood is
![\[ \log {(p(y))} = \sum _{i=1}^{n^{(3)}} w_ i \log {(p(y_ i))} \]](images/statug_glimmix0702.png)
where 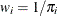 .
.
To illustrate weighting in a multilevel model, consider the following data set. In these simulated data, the response y is a Poisson-distributed count, w3 is the weight for the first-stage clusters, w2 is the weight for the second-stage clusters, and w1 is the weight for the observation-level units.
data d; input A w3 AB w2 w1 y x; datalines; 1 6.1 1 5.3 7.1 56 -.214 1 6.1 1 5.3 3.9 41 0.732 1 6.1 2 7.3 6.3 50 0.372 1 6.1 2 7.3 3.9 36 -.892 1 6.1 3 4.6 8.4 39 0.424 1 6.1 3 4.6 6.3 35 -.200 2 8.5 1 4.8 7.4 30 0.868 2 8.5 1 4.8 6.7 25 0.110 2 8.5 2 8.4 3.5 36 0.004 2 8.5 2 8.4 4.1 28 0.755 2 8.5 3 .80 3.8 33 -.600 2 8.5 3 .80 7.4 30 -.525 3 9.5 1 8.2 6.7 32 -.454 3 9.5 1 8.2 7.1 24 0.458 3 9.5 2 11 4.8 31 0.162 3 9.5 2 11 7.9 27 1.099 3 9.5 3 3.9 3.8 15 -1.57 3 9.5 3 3.9 5.5 19 -.448 4 4.5 1 8.0 5.7 30 -.468 4 4.5 1 8.0 2.9 25 0.890 4 4.5 2 6.0 5.0 35 0.635 4 4.5 2 6.0 3.0 30 0.743 4 4.5 3 6.8 7.3 17 -.015 4 4.5 3 6.8 3.1 18 -.560 ;
You can use the following statements to fit a weighted three-level model:
proc glimmix method=quadrature empirical=classical; class A AB; model y = x / dist=poisson link=log obsweight=w1; random int / subject=A weight=w3; random int / subject=AB(A) weight=w2; run;
The SUBJECT
= option in the first and second RANDOM
statements specifies the first-stage and second-stage clusters A and AB(A), respectively. The OBSWEIGHT
= option in the MODEL
statement specifies the variable for the weight at the observation level. The WEIGHT
= option in the RANDOM
statement specifies the variable for the weight at the level that is specified by the SUBJECT
= option.
For inference about fixed effects and variance that are estimated by pseudo-likelihood, you can use the empirical (sandwich) variance estimators. For weighted multilevel models, the only empirical estimator available in PROC GLIMMIX is EMPIRICAL =CLASSICAL. The EMPIRICAL =CLASSICAL variance estimator can be described as follows.
Let 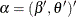 , where
, where 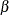 is vector of the fixed-effects parameters and
is vector of the fixed-effects parameters and 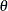 is the vector of covariance parameters. For an L-level model, Rabe-Hesketh and Skrondal (2006) show that the gradient can be written as a weighted sum of the gradients of the top-level units:
is the vector of covariance parameters. For an L-level model, Rabe-Hesketh and Skrondal (2006) show that the gradient can be written as a weighted sum of the gradients of the top-level units:
![\[ \sum _{i=1}^{n^{(L)}} w_ i \frac{\partial \log (p(y_{i};\alpha ))}{\partial \alpha } \equiv \sum _{i=1}^{n^{(L)}} S_ i(\alpha ) \]](images/statug_glimmix0706.png)
where 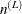 is the number of level-L units and
is the number of level-L units and 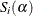 is the weighted score vector of the top-level unit i. The estimator of the "meat" of the sandwich estimator can be written as
is the weighted score vector of the top-level unit i. The estimator of the "meat" of the sandwich estimator can be written as
![\[ J= \frac{n^{(L)}}{n^{(L)}-1} \sum _{i=1}^{n^{(L)}} S_ i(\hat{\alpha })S_ i(\hat{\alpha })’ \]](images/statug_glimmix0709.png)
The empirical estimator of the covariance matrix of 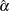 can be constructed as
can be constructed as
![\[ H(\hat{\alpha })^{-1}JH(\hat{\alpha })^{-1} \]](images/statug_glimmix0711.png)
where 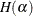 is the second derivative matrix of the log pseudo-likelihood with respect to
is the second derivative matrix of the log pseudo-likelihood with respect to  .
.
The covariance parameter estimators that are obtained by the pseudo-maximum-likelihood method can be biased when the sample
size is small. Pfeffermann et al. (1998) and Rabe-Hesketh and Skrondal (2006) discuss two weight-scaling methods for reducing the biases of the covariance parameter estimators in a two-level model.
To derive the scaling factor 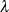 for a two-level model, let
for a two-level model, let 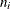 denote the number of level-1 units in the level-2 unit i and let
denote the number of level-1 units in the level-2 unit i and let 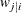 denote the weight of the jth level-1 unit in level-2 unit i. The first method computes an "apparent" cluster size as the "effective" sample size:
denote the weight of the jth level-1 unit in level-2 unit i. The first method computes an "apparent" cluster size as the "effective" sample size:
![\[ \sum _{j=1}^{n_ i} \lambda w_{j|i} = \frac{(\sum _{j=1}^{n_ i} w_{j|i})^2}{\sum _{j=1}^{n_ i} w^2_{j|i} } \]](images/statug_glimmix0714.png)
Therefore the scale factor is
![\[ \lambda = \frac{\sum _{j=1}^{n_ i} w_{j|i}}{\sum _{j=1}^{n_ i} w^2_{j|i}} \]](images/statug_glimmix0715.png)
The second method sets the apparent cluster size equal to the actual cluster size so that the scale factor is
![\[ \lambda =\frac{n_ i}{\sum _{j=1}^{n_ i} w_{j|i}} \]](images/statug_glimmix0716.png)
PROC GLIMMIX uses the weights provided in the data set directly. To use the scaled weights, you need to provide them in the data set.