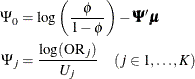The POWER Procedure
- Overview
-
Getting Started

-
Syntax

-
Details

-
Examples
 One-Way ANOVAThe Sawtooth Power Function in Proportion AnalysesSimple AB/BA Crossover DesignsNoninferiority Test with Lognormal DataMultiple Regression and CorrelationComparing Two Survival CurvesConfidence Interval PrecisionCustomizing PlotsBinary Logistic Regression with Independent PredictorsWilcoxon-Mann-Whitney Test
One-Way ANOVAThe Sawtooth Power Function in Proportion AnalysesSimple AB/BA Crossover DesignsNoninferiority Test with Lognormal DataMultiple Regression and CorrelationComparing Two Survival CurvesConfidence Interval PrecisionCustomizing PlotsBinary Logistic Regression with Independent PredictorsWilcoxon-Mann-Whitney Test - References
The power computing formula is based on Shieh and O’Brien (1998); Shieh (2000); Self, Mauritsen, and Ohara (1992), and Hsieh (1989).
Define the following notation for a logistic regression analysis:

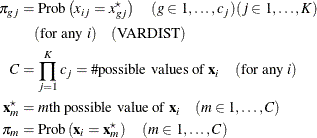
The logistic regression model is
The hypothesis test of the first predictor variable is
Assuming independence among all predictor variables, ![]() is defined as follows:
is defined as follows:
where ![]() is calculated according to the following algorithm:
is calculated according to the following algorithm:
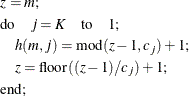
This algorithm causes the elements of the transposed vector ![]() to vary fastest to slowest from right to left as m increases, as shown in the following table of
to vary fastest to slowest from right to left as m increases, as shown in the following table of ![]() values:
values:
![\[ \begin{array}{cc|ccccc}& & & & j & & \\ h(m,j) & & 1 & 2 & \cdots & K-1 & K \\ \hline & 1 & 1 & 1 & \cdots & 1 & 1 \\ & 1 & 1 & 1 & \cdots & 1 & 2 \\ & \vdots & & & \vdots & & \\ & \vdots & 1 & 1 & \cdots & 1 & c_ K \\ & \vdots & 1 & 1 & \cdots & 2 & 1 \\ & \vdots & 1 & 1 & \cdots & 2 & 2 \\ & \vdots & & & \vdots & & \\ m & \vdots & 1 & 1 & \cdots & 2 & c_ K \\ & \vdots & & & \vdots & & \\ & \vdots & c_1 & c_2 & \cdots & c_{K-1} & 1 \\ & \vdots & c_1 & c_2 & \cdots & c_{K-1} & 2 \\ & \vdots & & & \vdots & & \\ & C & c_1 & c_2 & \cdots & c_{K-1} & c_ K \\ \end{array} \]](images/statug_power0091.png)
The ![]() values are determined in a completely analogous manner.
values are determined in a completely analogous manner.
The discretization is handled as follows (unless the distribution is ordinal, or binomial with sample size parameter at least
as large as requested number of bins): for ![]() , generate
, generate ![]() quantiles at evenly spaced probability values such that each such quantile is at the midpoint of a bin with probability
quantiles at evenly spaced probability values such that each such quantile is at the midpoint of a bin with probability ![]() . In other words,
. In other words,
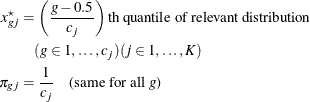
The primary noncentrality for the power computation is
where
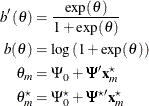
where
The power is
The factor ![]() is the adjustment for correlation between the predictor that is being tested and other predictors, from Hsieh (1989).
is the adjustment for correlation between the predictor that is being tested and other predictors, from Hsieh (1989).
Alternative input parameterizations are handled by the following transformations: