Overview of SAS/Genetics Software
Statistical analyses of genetic data are now central to medicine, agriculture, evolutionary biology, and forensic science. The inherent variation in genetic data, together with the substantial increase in the scale of genetic data following the human genome project, has created a need for reliable computer software to perform these analyses. The procedures offered by SAS/Genetics and described here represent an initial response of SAS Institute to this need.
Although many of the statistical techniques used in the new procedures are standard, others have had to be developed to reflect the genetic nature of the data. All the procedures are designed to operate on data sets that have a familiar structure to geneticists, and that mirror those used in existing software. The syntax for these genetic analyses follows that familiar to SAS users, and the output can be tabular or graphical. The objective of the procedures is to bring the full power of SAS analyses to bear on the characterization of fundamental genetic parameters, and most importantly on the detection of associations between genetic markers and disease status.
Most of the analyses in SAS/Genetics are concerned with detecting patterns of covariation in genetic marker data. These data generally consist of pairs of discrete categories; this pairing derives from the underlying biology, namely the fact that complex organisms have pairs of chromosomes. Each marker refers to the genetic status of a locus, each marker type is called an allele, and each pair of alleles in an individual is called a genotype. A set of alleles present on a single chromosome is called a haplotype. Genetic markers can be single nucleotide polymorphisms (SNPs), which are sites in the DNA where the nucleotide varies among individuals, usually with only two alleles possible; microsatellites, which are simple sequence repeats that generate usually between 2 and 20 categories; and other classes of DNA variation.
Two of the procedures in SAS/Genetics are concerned solely with the analysis of genetic marker data. The ALLELE procedure calculates descriptive statistics such as the frequency and variance of alleles and genotypes, as well as estimating measures of marker informativeness, and testing whether genotype frequencies are consistent with Hardy-Weinberg equilibrium (HWE). This procedure also supports four methods for calculation of the degree and significance of linkage disequilibrium (LD) among markers at pairs of loci, where LD refers to the propensity of alleles to co-segregate. The HAPLOTYPE procedure is used to infer the most likely multilocus haplotype frequencies in a set of genotypes. Since genetic markers are usually measured independently of one another, there is no direct way to determine which two alleles were on the same chromosome. The algorithm implemented in this procedure converges on the haplotype frequencies that have the highest probability of generating the observed genotypes. These estimated haplotype frequencies can be used as inputs to the HTSNP procedure where haplotype-tagging SNPs (htSNPs) that explain much of the haplotype diversity in a block or region can be identified.
Many genetic data sets are now used to study the relationship between genetic markers and complex phenotypes, particularly disease susceptibility. In general terms, traits can be measured as continuous variables (for example, weight or serum glucose concentration), as discrete numerical categories (for example, meristic measures or psychological class), or as affected/unaffected indicator variables. The two procedures CASECONTROL and FAMILY both take simple dichotomous indicators of disease status and use standard algorithms to compute statistics of association between these indicators and the genetic markers. The CASECONTROL procedure is designed to contrast allele and genotype frequencies between affected and unaffected populations, using three types of chi-square tests and options for controlling correlation of allele frequencies among members of the same subpopulation. Significant associations can indicate that the marker is linked to a locus that contributes to disease susceptibility, though population structure in conjunction with environmental or cultural variables can also lead to associations, and the statistical results must be interpreted with caution. The FAMILY procedure employs several transmission/disequilibrium tests of nonrandom association between disease status and linkage to markers transmitted from heterozygous parents to affected offspring (TDT) or pairs of affected and unaffected siblings (S-TDT and SDT). A joint analysis known as the reconstruction-combined TDT (RC-TDT) can also accommodate missing parental genotypes and families lacking unaffected children under some circumstances.
The output of these procedures can be further explored by using the PSMOOTH procedure to adjust 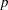 -values from association tests performed on large numbers of markers obtained in a genome scan, or by creating a graphical representation of the procedures’ output, namely
-values from association tests performed on large numbers of markers obtained in a genome scan, or by creating a graphical representation of the procedures’ output, namely 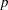 -values from tests for LD, HWE, and marker-disease associations, using the %TPLOT macro.
-values from tests for LD, HWE, and marker-disease associations, using the %TPLOT macro.
In addition to testing for associations between traits and single markers, testing for effects of multiple markers or epistatic effects between two or more markers might be of interest. PROC HAPLOTYPE can perform testing between a binary trait and several markers at the haplotype level, either looking across all haplotypes at a set of markers or testing each possible haplotype separately. The GENESELECT procedure can determine the best subset of markers, phenotypic variables, and their interactions to include in a model for predicting a trait.