| The ALLELE Procedure |
| Displayed Output |
This section describes the displayed output from PROC ALLELE. See the section ODS Table Names for details about how this output interfaces with the Output Delivery System.
Marker Summary
The "Marker Summary" table lists information about each of the markers, including the following:
NIndiv, the number of individuals genotyped at the marker
NAllele, the number of alleles at the marker
PIC, the polymorphism information content (PIC) measure
Het, the heterozygosity measure
Div, the allelic diversity measure
The table also contains the following columns for the test for HWE:
ChiSq, the chi-square statistic
DF, the degrees of freedom for the chi-square test
ProbChiSq, the
 -value for the chi-square test
-value for the chi-square test ProbExact, an estimate of the exact
 -value for the HWE test (only if the PERMS= option is specified in the PROC ALLELE statement)
-value for the HWE test (only if the PERMS= option is specified in the PROC ALLELE statement)
Allele Frequencies
The "Allele Frequencies" table lists all the observed alleles for each marker, with the observed allele count and frequency, the standard error of the frequency, and when the BOOTSTRAP= option is specified, the bootstrap lower and upper limits of the confidence interval for the frequency based on the confidence level determined by the ALPHA= option of the PROC ALLELE statement (0.95 by default).
Genotype Frequencies
The "Genotype Frequencies" table lists all the observed genotypes (denoted by the two alleles separated by a "/") for each marker, with the observed genotype count and frequency, an estimate of the disequilibrium coefficient 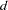 , the standard error of the estimate, and when the BOOTSTRAP= option is specified, the lower and upper limits of the bootstrap confidence interval for
, the standard error of the estimate, and when the BOOTSTRAP= option is specified, the lower and upper limits of the bootstrap confidence interval for 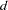 based on the confidence level determined by the ALPHA= option of the PROC ALLELE statement (0.95 by default).
based on the confidence level determined by the ALPHA= option of the PROC ALLELE statement (0.95 by default).
Linkage Disequilibrium Measures
The "Linkage Disequilibrium Measures" table lists for each marker pair the number of individuals with nonmissing genotypes, frequency of each haplotype (observed frequency when HAPLO=GIVEN and estimated frequency otherwise), an estimate of the LD coefficient 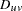 , and the linkage disequilibrium measures corresponding to the options included in the PROC ALLELE statement (CORRCOEFF, DELTA, DPRIME, PROPDIFF, RHO, and YULESQ). Haplotypes are represented by the allele at the marker locus listed in Locus1 and the allele at the marker locus listed in Locus2, separated by a "-." Note that this table can be quite large when there are many markers or markers with many alleles. For a data set with
, and the linkage disequilibrium measures corresponding to the options included in the PROC ALLELE statement (CORRCOEFF, DELTA, DPRIME, PROPDIFF, RHO, and YULESQ). Haplotypes are represented by the allele at the marker locus listed in Locus1 and the allele at the marker locus listed in Locus2, separated by a "-." Note that this table can be quite large when there are many markers or markers with many alleles. For a data set with 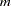 markers, each having
markers, each having 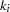 alleles,
alleles, 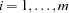 , the number of rows in the table is
, the number of rows in the table is 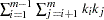 . The MAXDIST= option of the PROC ALLELE statement or the WITH statement can be used to keep this table to a manageable size.
. The MAXDIST= option of the PROC ALLELE statement or the WITH statement can be used to keep this table to a manageable size.
Population Summary
When the POP statement is specified, the "Population Summary" table displays the number and names of populations defined by the population variable.
Combined F Statistics
When the POP statement is specified, the "Combined 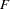 Statistics" table displays the estimated values of the following statistics combined over all marker loci:
Statistics" table displays the estimated values of the following statistics combined over all marker loci:
WithinPopf and OverallF, the inbreeding coefficients within populations and over all populations, respectively, when the ZEROF option is omitted
PopTheta, the degree of relatedness between individuals within populations
When the value specified for FPERMS is greater than 0, corresponding  -values for each of these estimates are also reported.
-values for each of these estimates are also reported.
Marker F Statistics
When the POP statement is specified with the INDIVLOCI option, the "Marker 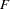 Statistics" table displays the same columns as those in the "Combined
Statistics" table displays the same columns as those in the "Combined 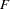 Statistics" table for each of the marker variables represented in the VAR statement.
Statistics" table for each of the marker variables represented in the VAR statement.
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.