The MODEL Procedure
Example 26.22 Reducing Parameter Variance in a Tree Biomass Model
This example uses various dimensions of willow oak trees to model their biomass. Unlike a tree’s biomass, a tree’s dimensions can be measured noninvasively. The model and data for this example are taken from Parresol (1999). This biomass model uses four equations to model the bole wood, bole bark, crown, and total mass of the trees,
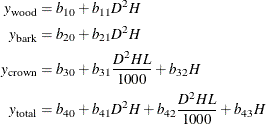
where 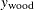 ,
, 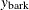 , and
, and 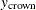 are the three components of a tree’s total biomass,
are the three components of a tree’s total biomass, 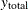 ; D is the tree’s diameter at breast height; H is the tree’s height; and L is the tree’s live crown length. The efficiency of parameter estimates in this model is improved by taking into account the
correlations between errors in the equations for the four components of the trees’ biomass. Also, the efficiency of estimates
is improved by taking into account heteroscedasticity in the sample of 39 trees that is used to develop the model.
; D is the tree’s diameter at breast height; H is the tree’s height; and L is the tree’s live crown length. The efficiency of parameter estimates in this model is improved by taking into account the
correlations between errors in the equations for the four components of the trees’ biomass. Also, the efficiency of estimates
is improved by taking into account heteroscedasticity in the sample of 39 trees that is used to develop the model.
In an initial OLS estimation of this model’s parameters, some general properties of the model and data can be quantified using the following statements:
proc model data=trees; endo wood bark crown total; vol = dbh**2*height; cvol = vol*lcl/1000; wood = b10 + b11*vol; bark = b20 + b21*vol; crown = b30 + b31*cvol + b32*height; total = b40 + b41*vol + b42*cvol + b43*height; fit / ols outs=stree out=otree outresid; quit;
Here the covariance of equation errors is saved to the data set STREE, and the residuals are saved to the data set OTREE for later analysis.
The covariance matrix of equation errors that is computed in the OLS estimation can be used in a seemingly unrelated regression (SUR) estimation of the model to improve the efficiency of parameter estimates. The total biomass of trees is restricted to equal the other three biomass components in the following SUR estimation:
proc model data=trees;
endo wood bark crown total;
vol = dbh**2*height;
cvol = vol*lcl/1000;
wood = b10 + b11*vol;
bark = b20 + b21*vol;
crown = b30 + b31*cvol + b32*height;
total = b40 + b41*vol + b42*cvol + b43*height;
fit / nools sur sdata=stree;
restrict b10 + b20 + b30 = b40,
b11 + b21 = b41,
b42 = b31,
b43 = b32;
quit;
Output 26.22.1 shows the parameters and standard errors for the restricted SUR tree biomass model.
Output 26.22.1: SUR Parameter Estimates for Willow Oak Model
| Nonlinear SUR Parameter Estimates | |||||
|---|---|---|---|---|---|
| Parameter | Estimate | Approx Std Err | t Value | Approx Pr > |t| |
Label |
| b10 | 59.18653 | 48.2435 | 1.23 | 0.2274 | |
| b11 | 0.026598 | 0.000558 | 47.67 | <.0001 | |
| b20 | 29.65101 | 10.3839 | 2.86 | 0.0069 | |
| b21 | 0.003225 | 0.000120 | 26.79 | <.0001 | |
| b30 | 133.1066 | 26.6246 | 5.00 | <.0001 | |
| b31 | 0.061543 | 0.00508 | 12.11 | <.0001 | |
| b32 | -5.33604 | 1.1141 | -4.79 | <.0001 | |
| b40 | 221.9441 | 62.1834 | 3.57 | 0.0010 | |
| b41 | 0.029824 | 0.000623 | 47.91 | <.0001 | |
| b42 | 0.061543 | 0.00508 | 12.11 | <.0001 | |
| b43 | -5.33604 | 1.1141 | -4.79 | <.0001 | |
| Restrict0 | -656E-14 | 0.1316 | -0.00 | 1.0000 | b10 + b20 + b30 = b40 |
| Restrict1 | 148.5607 | 11355.0 | 0.01 | 0.9898 | b11 + b21 = b41 |
| Restrict2 | 68.66564 | 178.8 | 0.38 | 0.7067 | b42 = b31 |
| Restrict3 | 0.388714 | 3.5449 | 0.11 | 0.9145 | b43 = b32 |
An analysis of heteroscedasticity in the unrestricted OLS model suggests that the efficiency of the estimation could be improved further by weighting the observations using the following variance model:
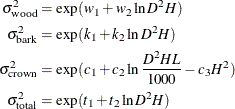
where 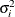 is the variance of the ith component of the biomass and the parameters
is the variance of the ith component of the biomass and the parameters 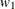 ,
, 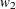 ,
, 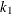 ,
, 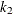 ,
, 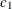 ,
, 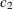 ,
, 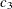 ,
, 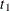 , and
, and 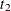 are determined by regressing the square of the residuals from the unrestricted OLS estimation against the tree dimensions.
Estimates of the parameters in the variance model can be determined using the following statements:
are determined by regressing the square of the residuals from the unrestricted OLS estimation against the tree dimensions.
Estimates of the parameters in the variance model can be determined using the following statements:
proc model data=otree; vol = dbh**2*height; cvol = vol*lcl/1000; eq.varwood = log(wood*wood) - (w1 + w2*log(vol)); eq.varbark = log(bark*bark) - (k1 + k2*log(vol)); eq.varcrown = log(crown*crown) - (c1 + c2*log(cvol) - c3*height**2); eq.vartotal = log(total*total) - (t1 + t2*log(vol)); fit varwood varbark varcrown vartotal; quit;
The biomass component variance model can be used to account for heteroscedasticity and improve the efficiency of parameter estimates by weighting observations in the biomass model. The following PROC MODEL statements accommodate both the covariance among the biomass equations’ errors and the heteroscedasticity that is observed in each component of the trees’ biomass:
proc model data=trees;
endo wood bark crown total;
vol = dbh**2*height;
cvol = vol*lcl/1000;
wood = b10 + b11*vol;
bark = b20 + b21*vol;
crown = b30 + b31*cvol + b32*height;
total = b40 + b41*vol + b42*cvol + b43*height;
h.wood = exp(&w1)*vol**&w2;
h.bark = exp(&k1)*vol**&k2;
h.crown = exp(&c1)*cvol**&c2 * exp (-&c3*height*height);
h.total = exp(&t1)*vol**&t2;
fit / sur;
restrict b10 + b20 + b30 = b40,
b11 + b21 = b41,
b42 = b31,
b43 = b32;
quit;
Output 26.22.2 shows the parameters and standard errors for the heteroscedastic tree biomass model.
Output 26.22.2: SUR Parameter Estimates for Heteroscedastic Willow Oak Model
| Nonlinear SUR Parameter Estimates | |||||
|---|---|---|---|---|---|
| Parameter | Estimate | Approx Std Err | t Value | Approx Pr > |t| |
Label |
| b10 | 10.22253 | 14.2208 | 0.72 | 0.4766 | |
| b11 | 0.027515 | 0.000534 | 51.48 | <.0001 | |
| b20 | 15.66358 | 4.8173 | 3.25 | 0.0024 | |
| b21 | 0.003476 | 0.000138 | 25.13 | <.0001 | |
| b30 | 103.8782 | 10.6122 | 9.79 | <.0001 | |
| b31 | 0.055226 | 0.00306 | 18.02 | <.0001 | |
| b32 | -4.05163 | 0.4603 | -8.80 | <.0001 | |
| b40 | 129.7643 | 21.5621 | 6.02 | <.0001 | |
| b41 | 0.030991 | 0.000614 | 50.47 | <.0001 | |
| b42 | 0.055226 | 0.00306 | 18.02 | <.0001 | |
| b43 | -4.05163 | 0.4603 | -8.80 | <.0001 | |
| Restrict0 | -0.28369 | 1.1969 | -0.24 | 0.8164 | b10 + b20 + b30 = b40 |
| Restrict1 | -56046.2 | 49194.0 | -1.14 | 0.2603 | b11 + b21 = b41 |
| Restrict2 | 539.3854 | 697.4 | 0.77 | 0.4471 | b42 = b31 |
| Restrict3 | 4.374001 | 29.6959 | 0.15 | 0.8853 | b43 = b32 |
Output 26.22.3 shows the improved efficiency of estimates in the heteroscedastic model as compared to the homoscedastic model. For most of the parameters, the standard errors are reduced significantly in the heteroscedastic estimation.
Output 26.22.3: Standard Error Reduction