The MODEL Procedure
ODS Table Names
PROC MODEL assigns a name to each table it creates. You can use these names to reference the table when you use the Output Delivery System (ODS) to select tables and create output data sets. These names are listed in the following table.
Table 26.3: ODS Tables Produced in PROC MODEL
|
ODS Table Name |
Description |
Option |
|---|---|---|
|
ODS Tables Created by the FIT Statement |
||
|
AugGMMCovariance |
Crossproducts matrix |
GMM ITALL |
|
ChowTest |
Structural change test |
CHOW= |
|
CollinDiagnostics |
Collinearity diagnostics |
|
|
ConfInterval |
Profile likelihood confidence intervals |
PRL= |
|
ConvCrit |
Convergence criteria for estimation |
default |
|
ConvergenceStatus |
Convergence status |
default |
|
CorrB |
Correlations of parameters |
COVB/CORRB |
|
CorrResiduals |
Correlations of residuals |
CORRS/COVS |
|
CovB |
Covariance of parameters |
COVB/CORRB |
|
CovResiduals |
Covariance of residuals |
CORRS/COVS |
|
Crossproducts |
Crossproducts matrix |
ITALL/ITPRINT |
|
DatasetOptions |
Data sets used |
default |
|
DetResidCov |
Determinant of the residuals |
DETAILS |
|
DWTest |
Durbin Watson test |
DW= |
|
Equations |
Listing of equations to estimate |
default |
|
EstSummaryMiss |
Model summary statistics for PAIRWISE |
MISSING= |
|
EstSummaryStats |
Objective, objective * N |
default |
|
FirstLagrMultEst |
First order Lagrange Multiplier estimates |
GMM ITALL |
|
GMMCovariance |
Crossproducts matrix |
GMM DETAILS |
|
GMMTestStats |
GMM test statistics |
GMM |
|
Godfrey |
Godfrey’s serial correlation test |
GF= |
|
HausmanTest |
Hausman’s test table |
HAUSMAN |
|
HeteroTest |
Heteroscedasticity test tables |
BREUSCH/PAGEN |
|
InvXPXMat |
X |
I |
|
IterInfo |
Iteration printing |
ITALL/ITPRINT |
|
LagLength |
Model lag length |
default |
|
MinSummary |
Number of parameters, estimation kind |
default |
|
ModSummary |
Listing of all categorized variables |
default |
|
ModVars |
Listing of model variables and parameters |
default |
|
NormalityTest |
Normality test table |
NORMAL |
|
ObsSummary |
Identifies observations with errors |
default |
|
ObsUsed |
Observations read, used, and missing. |
default |
|
ParameterEstimates |
Parameter estimates |
default |
|
ParmChange |
Parameter change vector |
ITALL |
|
ResidSummary |
Summary of the SSE, MSE for the equations |
default |
|
SecondLagrMultEst |
Second order Lagrange Multiplier estimates |
GMM ITALL |
|
SizeInfo |
Storage requirement for estimation |
DETAILS |
|
TermEstimates |
Nonlinear OLS and ITOLS Estimates |
OLS/ITOLS |
|
TestResults |
Test statement table |
|
|
WgtVar |
The name of the weight variable |
|
|
XPXMat |
X |
XPX |
|
YkVector |
Marquardt iteration vector |
GMM ITALL |
|
ODS Tables Created by the SOLVE Statement |
||
|
BlockEqsAndVars |
Dependency analysis block partitioning |
ANALYZEDEPS= |
|
DatasetOptions |
Data sets used |
default |
|
DescriptiveStatistics |
Descriptive statistics |
STATS |
|
FitStatistics |
Fit statistics for simulation |
STATS |
|
LagLength |
Model lag length |
default |
|
ModSummary |
Listing of all categorized variables |
default |
|
ObsSummary |
Simulation trace output |
SOLVEPRINT |
|
ObsUsed |
Observations read, used, and missing. |
default |
|
SimulationSummary |
Number of variables solved for |
default |
|
SolutionVarList |
Solution variable lists |
default |
|
TheilRelStats |
Theil relative change error statistics |
THEIL |
|
TheilStats |
Theil forecast error statistics |
THEIL |
|
ErrorVec |
Iteration Error vector |
ITPRINT |
|
ResidualValues |
Iteration residual values |
ITPRINT |
|
PredictedValues |
Iteration predicted values |
ITPRINT |
|
SolutionValues |
Iteration solved for variable values |
ITPRINT |
|
ODS Tables Created by the FIT and SOLVE Statements |
||
|
AdjacencyMatrix |
Adjacency graph |
GRAPH |
|
BlockAnalysis |
Block analysis |
BLOCK |
|
BlockStructure |
Block structure |
BLOCK |
|
CodeDependency |
Variable cross reference |
LISTDEP |
|
CodeList |
Listing of programs statements |
LISTCODE |
|
CrossReference |
Cross-reference listing for program |
|
|
DepStructure |
Dependency structure of the system |
BLOCK |
|
FirstDerivatives |
First derivative table |
LISTDER |
|
IterIntg |
Integration iteration output |
INTGPRINT |
|
MemUsage |
Memory usage statistics |
MEMORYUSE |
|
MissingDependencies |
Missing values by dependency |
REPORTMISSINGS |
|
MissingObservations |
Missing values by observation |
REPORTMISSINGS |
|
MissingSymbols |
Missing values by symbol |
REPORTMISSINGS |
|
ParmReadIn |
Parameter estimates read in |
ESTDATA= |
|
ProgList |
Listing of compiled program code |
|
|
RangeInfo |
RANGE statement specification |
|
|
SortAdjacencyMatrix |
Sorted adjacency graph |
GRAPH |
|
TransitiveClosure |
Transitive closure graph |
GRAPH |
The AugGMMCovariance table is the 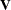 matrix augmented with the moment vector at iteration zero, produced when the ITALL option is used with the GMM option. If
the
matrix augmented with the moment vector at iteration zero, produced when the ITALL option is used with the GMM option. If
the 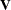 matrix to be used in GMM is read in by the VDATA option, then AugGMMCovariance would be the same matrix augmented with the
moment vectors. The GMMCovariance ODS output is produced only when you read in a covariance matrix to be used in the GMM method.
This table is produced by using DETAILS option with the GMM option.
matrix to be used in GMM is read in by the VDATA option, then AugGMMCovariance would be the same matrix augmented with the
moment vectors. The GMMCovariance ODS output is produced only when you read in a covariance matrix to be used in the GMM method.
This table is produced by using DETAILS option with the GMM option.
