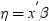Generalized Linear Models
About the Generalized Linear Models Task
The Generalized Linear
Models task is a high-performance task that provides model fitting
and model building for generalized linear models. It fits models for
standard distributions such as normal, Poisson, and Tweedie in the
exponential family. This task also fits multinomial models for ordinal
and nominal responses. The task provides forward, backward, and stepwise
selection methods.
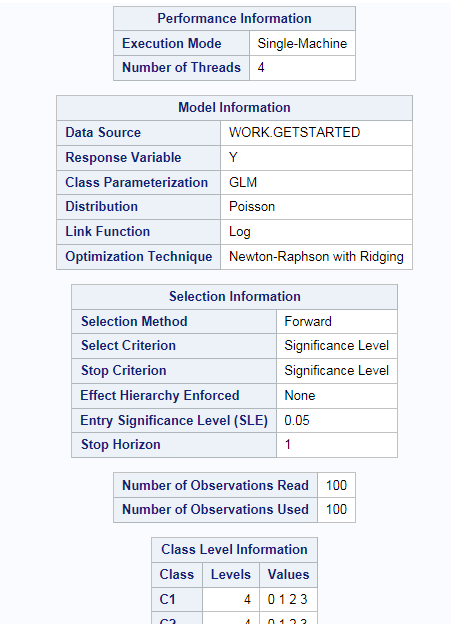
Example: Model Selection
To create this example:
-
Create the Work.getStarted data set. For more information, see GETSTARTED Data Set.
Assigning Data to Roles
To run the Generalized
Linear Model task, you must assign a column to the Response
variable role.
Building a Model
Requirements for Building a Model
By default, no effects
are specified, which results in the task fitting an intercept-only
model. To specify an effect, you must assign at least one variable
to the Continuous variables or Classification
variables role. You can select combinations of variables
to create crossed, factorial, or polynomial effects.
Setting the Model Options
Copyright © SAS Institute Inc. All rights reserved.