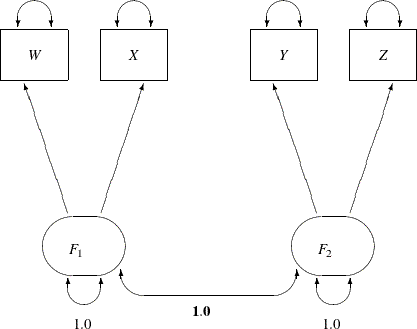
The path diagram for this congeneric (one-factor) model is shown in Figure 17.20.
The only difference between the current path diagram in Figure 17.20 for the congeneric (one-factor) model and the preceding path diagram in Figure 17.17 for the full (two-factor) model is that the double-headed path that connects F1 and F2 is fixed to 1 in the current path diagram. Accordingly, you need to modify only slightly the preceding PROC CALIS specification
to form the new model specification, as shown in the following statements:
proc calis data=lord;
path
W <=== F1,
X <=== F1,
Y <=== F2,
Z <=== F2;
pvar
F1 = 1.0,
F2 = 1.0,
W X Y Z;
pcov
F1 F2 = 1.0;
run;
This specification sets the covariance between F1 and F2 to 1.0 in the PCOV statement. An annotated fit summary is displayed in Figure 17.21.
Figure 17.21: Fit Summary, H3: Congeneric (One-Factor) Model for Lord Data
| Fit Summary | |
|---|---|
| Chi-Square | 36.2095 |
| Chi-Square DF | 2 |
| Pr > Chi-Square | <.0001 |
| Standardized RMR (SRMR) | 0.0277 |
| Adjusted GFI (AGFI) | 0.8570 |
| RMSEA Estimate | 0.1625 |
| Bentler Comparative Fit Index | 0.9766 |
The chi-square value is 36.2095 (df = 2, p < 0.0001). This indicates that you can reject the hypothesized model at the 0.01 ![]() -level. The standardized root mean square error (SRMSR) is 0.0277, which indicates a good fit. Bentler’s comparative fit index
is 0.9766, which is also a good model fit. However, the adjusted GFI (AGFI) is 0.8570, which is not very impressive. Also,
the RMSEA value is 0.1625, which is too large to be an acceptable model. Therefore, the congeneric model might not be the
one you want to use.
-level. The standardized root mean square error (SRMSR) is 0.0277, which indicates a good fit. Bentler’s comparative fit index
is 0.9766, which is also a good model fit. However, the adjusted GFI (AGFI) is 0.8570, which is not very impressive. Also,
the RMSEA value is 0.1625, which is too large to be an acceptable model. Therefore, the congeneric model might not be the
one you want to use.
The estimation results are displayed in Figure 17.22. Because the model does not fit well, the corresponding estimation results are not interpreted.
Figure 17.22: Estimation Results, H3: Congeneric (One-Factor) Model for Lord Data
| PATH List | ||||||
|---|---|---|---|---|---|---|
| Path | Parameter | Estimate | Standard Error |
t Value | ||
| W | <=== | F1 | _Parm1 | 7.10470 | 0.32177 | 22.08012 |
| X | <=== | F1 | _Parm2 | 7.26908 | 0.31826 | 22.83973 |
| Y | <=== | F2 | _Parm3 | 8.37344 | 0.32542 | 25.73143 |
| Z | <=== | F2 | _Parm4 | 8.51060 | 0.32409 | 26.26002 |
| Variance Parameters | |||||
|---|---|---|---|---|---|
| Variance Type |
Variable | Parameter | Estimate | Standard Error |
t Value |
| Exogenous | F1 | 1.00000 | |||
| F2 | 1.00000 | ||||
| Error | W | _Parm5 | 35.92111 | 2.41467 | 14.87619 |
| X | _Parm6 | 33.42373 | 2.31037 | 14.46684 | |
| Y | _Parm7 | 27.17043 | 2.24621 | 12.09613 | |
| Z | _Parm8 | 25.38887 | 2.20837 | 11.49664 | |
| Covariances Among Exogenous Variables | ||||
|---|---|---|---|---|
| Var1 | Var2 | Estimate | Standard Error |
t Value |
| F1 | F2 | 1.00000 | ||
Perhaps a more natural way to specify the model under hypothesis ![]() is to use only one factor in the PATH model, as shown in the following statements:
is to use only one factor in the PATH model, as shown in the following statements:
proc calis data=lord;
path
W <=== F1,
X <=== F1,
Y <=== F1,
Z <=== F1;
pvar
F1 = 1.0,
W X Y Z;
run;
This produces essentially the same results as the specification with two factors that have perfect correlation.