| The GENMOD Procedure |
| Displayed Output for Bayesian Analysis |
If a Bayesian analysis is requested with a BAYES statement, the displayed output includes the following.
Model Information
The "Model Information" table displays the two-level data set name, the number of burn-in iterations, the number of iterations after the burn-in, the number of thinning iterations, the response distribution, the link function, the response variable name, the offset variable name, the frequency variable name, the scale weight variable name, the number of observations used, the number of events if events/trials format is used for response, the number of trials if events/trials format is used for response, the sum of frequency weights, the number of missing values in data set, and the number of invalid observations (for example, negative or 0 response values with gamma distribution or number of observations with events greater than trials with binomial distribution).
Class Level Information
The "Class Level Information" table displays the levels of classification variables if you specify a CLASS statement.
Maximum Likelihood Estimates
The "Analysis of Maximum Likelihood Parameter Estimates" table displays the maximum likelihood estimate of each parameter, the estimated standard error of the parameter estimator, and confidence limits for each parameter.
Coefficient Prior
The "Coefficient Prior" table displays the prior distribution of the regression coefficients.
Independent Prior Distributions for Model Parameters
The "Independent Prior Distributions for Model Parameters" table displays the prior distributions of additional model parameters (scale, exponential scale, Weibull scale, Weibull shape, gamma shape).
Initial Values and Seeds
The "Initial Values and Seeds" table displays the initial values and random number generator seeds for the Gibbs chains.
Fit Statistics
The "Fit Statistics" table displays the deviance information criterion (DIC) and the effective number of parameters.
Descriptive Statistics of the Posterior Samples
The "Descriptive Statistics of the Posterior Sample" table contains the size of the sample, the mean, the standard deviation, and the quartiles for each model parameter.
Interval Estimates for Posterior Sample
The "Interval Estimates for Posterior Sample" table contains the HPD intervals and the credible intervals for each model parameter.
Correlation Matrix of the Posterior Samples
The "Correlation Matrix of the Posterior Samples" table is produced if you include the CORR suboption in the SUMMARY= option in the BAYES statement. This table displays the sample correlation of the posterior samples.
Covariance Matrix of the Posterior Samples
The "Covariance Matrix of the Posterior Samples" table is produced if you include the COV suboption in the SUMMARY= option in the BAYES statement. This table displays the sample covariance of the posterior samples.
Autocorrelations of the Posterior Samples
The "Autocorrelations of the Posterior Samples" table displays the lag1, lag5, lag10, and lag50 autocorrelations for each parameter.
Gelman and Rubin Diagnostics
The "Gelman and Rubin Diagnostics" table is produced if you include the GELMAN suboption in the DIAGNOSTIC= option in the BAYES statement. This table displays the estimate of the potential scale reduction factor and its 97.5% upper confidence limit for each parameter.
Geweke Diagnostics
The "Geweke Diagnostics" table displays the Geweke statistic and its 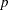 -value for each parameter.
-value for each parameter.
Raftery and Lewis Diagnostics
The "Raftery Diagnostics" tables is produced if you include the RAFTERY suboption in the DIAGNOSTIC= option in the BAYES statement. This table displays the Raftery and Lewis diagnostics for each variable.
Heidelberger and Welch Diagnostics
The "Heidelberger and Welch Diagnostics" table is displayed if you include the HEIDELBERGER suboption in the DIAGNOSTIC= option in the BAYES statement. This table shows the results of a stationary test and a halfwidth test for each parameter.
Effective Sample Size
The "Effective Sample Size" table displays, for each parameter, the effective sample size, the correlation time, and the efficiency.
Monte Carlo Standard Errors
The "Monte Carlo Standard Errors" table displays, for each parameter, the Monte Carlo standard error, the posterior sample standard deviation, and the ratio of the two.
Copyright © SAS Institute, Inc. All Rights Reserved.