| The GENMOD Procedure |
| ESTIMATE Statement |
- ESTIMATE ’label’ effect values <,...effect values> </options> ;
The ESTIMATE statement is similar to a CONTRAST statement, except only one-row 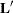 matrices are permitted.
matrices are permitted.
In the case of zero-inflated (ZI) models, the statement syntax is:
- ESTIMATE ’label’ effect values <,...effect values> @zero effect values <,...effect values> </options> ;
where sets of effects values before the @zero separator correspond to the regression part of the model, and effects values after the @zero separator correspond to the zero-inflation part of the model. In the case of ZI models, a one-row 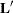 matrix is created for the regression part of the model, another one-row
matrix is created for the regression part of the model, another one-row 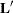 matrix is created for the zero-inflation part of the model, and separate estimates for the two
matrix is created for the zero-inflation part of the model, and separate estimates for the two 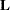 matrices are computed and displayed.
matrices are computed and displayed.
If you use the default less-than-full-rank GLM CLASS variable parameterization, each row is checked for estimability. If PROC GENMOD finds a contrast to be nonestimable, it displays missing values in corresponding rows in the results. See Searle (1971) for a discussion of estimable functions.
The actual estimate,  , its approximate standard error, and its confidence limits are displayed. A Wald chi-square test that
, its approximate standard error, and its confidence limits are displayed. A Wald chi-square test that  = 0 is also displayed.
= 0 is also displayed.
The approximate standard error of the estimate is computed as the square root of  , where
, where  is the estimated covariance matrix of the parameter estimates. If you specify a GEE model in the REPEATED statement,
is the estimated covariance matrix of the parameter estimates. If you specify a GEE model in the REPEATED statement,  is the empirical covariance matrix estimate.
is the empirical covariance matrix estimate.
If you specify the EXP option, then  , its standard error, and its confidence limits are also displayed.
, its standard error, and its confidence limits are also displayed.
The construction of the 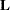 vector and the checking for estimability for an ESTIMATE statement follow the same rules as listed under the CONTRAST statement.
vector and the checking for estimability for an ESTIMATE statement follow the same rules as listed under the CONTRAST statement.
You can specify the following options in the ESTIMATE statement after a slash (/).
- ALPHA=number
requests that a confidence interval be constructed with confidence level
 . The value of number must be between 0 and 1; the default value is 0.05.
. The value of number must be between 0 and 1; the default value is 0.05. - E
- EXP
requests that
 , its standard error, and its confidence limits be computed. If you specify the EXP option, standard errors and confidence intervals are computed using the delta method.
, its standard error, and its confidence limits be computed. If you specify the EXP option, standard errors and confidence intervals are computed using the delta method. - SINGULAR=number
- EPSILON=number
tunes the estimability checking as described for the CONTRAST statement.
Copyright © SAS Institute, Inc. All Rights Reserved.