| What’s New in SAS/STAT |
GLIMMIX Procedure
The GLIMMIX procedure fits statistical models to data with correlations or nonconstant variability and where the response is not necessarily normally distributed. These generalized linear mixed models (GLMM), like linear mixed models, assume normal (Gaussian) random effects. Conditional on these random effects, data can have any distribution in the exponential family. The binary, binomial, Poisson, and negative binomial distributions, for example, are discrete members of this family. The normal, beta, gamma, and chi-square distributions are representatives of the continuous distributions in this family. The GLIMMIX procedure was first made available for SAS 9.1.3 as a Web download.
In SAS 9.2, the GLIMMIX procedure provides Laplace and adaptive quadrature estimation methods, and, with them, a likelihood-based empirical estimator. In addition, a new bias-corrected estimator is available. The experimental EFFECT statement provides for the creation of splines as well as other special effects. The COVTEST statement enables likelihood-based inference about the covariance parameters. A number of additional covariance structures have been added, including heterogeneous AR(1), heterogeneous compound symmetry, linear structures, heterogeneous Toeplitz, penalized B-spline, spatial anisotropic, and the Matérn covariance structure. Step-down multiplicity adjustments are now supported for all ADJUST= methods in the LSMEANS, ESTIMATE, and LSMESTIMATE statements, except for ADJUST=NELSON in the LSMEANS statement.
The DDFM=KR(FIRSTRORDER) option drops the second-derivative term in the KR calculations. The OUTDESIGN= option in the PROC GLIMMIX statement enables you to write the 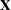 and
and 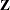 matrix to an output data set. New graphics include boxplots of data and/or residuals with respect to classification effects as well as plots of odds ratios and their confidence limits. The diffogram, meanplot, anomplot, and controlplot have been enhanced.
matrix to an output data set. New graphics include boxplots of data and/or residuals with respect to classification effects as well as plots of odds ratios and their confidence limits. The diffogram, meanplot, anomplot, and controlplot have been enhanced.
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.