General Statistics Examples
Example 10.5 Categorical Linear Models
This example fits a linear model to a function of the response probabilities
![\[ \mb{K} \log \pi = \mb{X \beta } + \epsilon \]](images/imlug_genstatexpls0009.png)
where 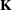 is a matrix that compares each response category to the last category.
is a matrix that compares each response category to the last category.
First, the Grizzle-Starmer-Koch approach (Grizzle, Starmer, and Koch 1969) is used to obtain generalized least squares estimates of 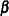 . These form the initial values for the Newton-Raphson solution for the maximum likelihood estimates. The CATMOD procedure
can also be used to analyze these binary data (Cox 1970).
. These form the initial values for the Newton-Raphson solution for the maximum likelihood estimates. The CATMOD procedure
can also be used to analyze these binary data (Cox 1970).
proc iml ;
/* Subroutine to compute new probability estimates */
/* Last column not needed since sum of each row is 1 */
start prob(x, beta, q);
la = exp(x*shape(beta,0,q));
pi = la / ((1+la[,+])*repeat(1,1,q));
return( colvec(pi) );
finish prob;
/* Categorical Linear Models */
/* by Least Squares and Maximum Likelihood */
/* Input: */
/* n the s by p matrix of response counts */
/* x the s by r design matrix */
start catlin(n, x);
s = nrow(n); /* number of populations */
r = ncol(n); /* number of responses */
q = r-1; /* number of function values */
d = ncol(x); /* number of design parameters */
qd = q*d; /* total number of parameters */
/* initial (emprical) probability estimates */
rown = n[,+]; /* row totals */
pr = n/rown; /* probability estimates */
print pr[label="Initial Probability Estimates"];
/* function of probabilities */
p = colvec(pr[,1:q]); /* cut and shaped to vector */
f = log(p) - log(pr[,r])@repeat(1,q,1);
/* estimate by the GSK method */
/* inverse covariance of f */
si = (diag(p)-p*p`) # (diag(rown)@repeat(1,q,q));
z = x@I(q); /* expanded design matrix */
h = z`*si*z; /* crossproducts matrix */
g = z`*si*f; /* cross with f */
beta = solve(h,g); /* least squares solution */
stderr = sqrt(vecdiag(inv(h))); /* standard errors */
pi = prob(x, beta, q);
est = beta || stderr;
pr = shape(pi, 0, q);
print est[colname={"beta" "stderr"} label="GSK Estimates"], pr;
/* ML solution */
crit = 1;
do it = 1 to 8 while(crit>.0005);/* iterate until converge*/
/* block diagonal weighting */
si = (diag(pi)-pi*pi`) #( diag(rown)@repeat(1,q,q));
g = z`*(rown@repeat(1,q,1)#(p-pi)); /* gradient */
h = z`*si*z; /* hessian */
delta = solve(h,g); /* correction via Newton's method */
beta = beta+delta; /* apply the correction */
pi = prob(x, beta, q); /* compute prob estimates */
crit = max(abs(delta)); /* convergence criterion */
end;
stderr = sqrt(vecdiag(inv(h))); /* standard errors */
est = beta || stderr;
pr = shape(pi, 0, q);
print est[colname={"beta" "stderr"} label="ML Estimates"], pr;
print it[label="Iterations"] crit[label="Criterion"];
finish catlin;
The following statements call the CATLIN module to analyze data from Kastenbaum and Lamphiear (1959):
/* frequency counts*/
n= { 58 11 05,
75 19 07,
49 14 10,
58 17 08,
33 18 15,
45 22 10,
15 13 15,
39 22 18,
04 12 17,
05 15 08};
/* design matrix */
x= { 1 1 1 0 0 0,
1 -1 1 0 0 0,
1 1 0 1 0 0,
1 -1 0 1 0 0,
1 1 0 0 1 0,
1 -1 0 0 1 0,
1 1 0 0 0 1,
1 -1 0 0 0 1,
1 1 -1 -1 -1 -1,
1 -1 -1 -1 -1 -1};
run catlin(n, x);
The maximum likelihood estimates are shown in Output 10.5.1.
Output 10.5.1: Maximum Likelihood Estimates
| Initial Probability Estimates | ||
|---|---|---|
| 0.7837838 | 0.1486486 | 0.0675676 |
| 0.7425743 | 0.1881188 | 0.0693069 |
| 0.6712329 | 0.1917808 | 0.1369863 |
| 0.6987952 | 0.2048193 | 0.0963855 |
| 0.5 | 0.2727273 | 0.2272727 |
| 0.5844156 | 0.2857143 | 0.1298701 |
| 0.3488372 | 0.3023256 | 0.3488372 |
| 0.4936709 | 0.278481 | 0.2278481 |
| 0.1212121 | 0.3636364 | 0.5151515 |
| 0.1785714 | 0.5357143 | 0.2857143 |