The TSPLINE subroutine fits a thin-plate smoothing spline (TPSS) to data. The generalized cross validation (GCV) function is used to select the smoothing parameter.
The TPSPLINE subroutine returns the following values:
- fitted
-
is an
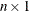 vector of fitted values of the TPSS fit evaluated at the design points
vector of fitted values of the TPSS fit evaluated at the design points  . The
. The  is the number of observations. The final TPSS fit depends on the optional lambda.
is the number of observations. The final TPSS fit depends on the optional lambda.
- coeff
-
is a vector of spline coefficients. The vector contains the coefficients for basis functions in the null space and the representer of evaluation functions at unique design points. (see Wahba (1990) for more detail on reproducing kernel Hilbert space and representer of evaluation functions.) The length of coeff vector depends on the number of unique design points and the number of variables in the spline model. In general, let nuobs and
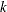 be the number of unique rows and the number of columns of
be the number of unique rows and the number of columns of  respectively. The length of coeff equals to
respectively. The length of coeff equals to 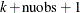 . The coeff vector can be used as an input to the TPSPLNEV subroutine to evaluate the resulting TPSS fit at new data points.
. The coeff vector can be used as an input to the TPSPLNEV subroutine to evaluate the resulting TPSS fit at new data points.
- adiag
-
is an
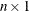 vector of diagonal elements of the “hat” matrix. See the “Details” section.
vector of diagonal elements of the “hat” matrix. See the “Details” section.
- gcv
-
If lambda is not specified, then gcv is the minimum value of the GCV function. If lambda is specified, then gcv is a vector (or scalar if lambda is a scalar) of GCV values evaluated at the lambda points. It provides you with both the ability to study the GCV curves by plotting gcv against lambda and the chance to identify a possible local minimum.
The input arguments to the TPSPLINE subroutine are as follows:
- x
-
is an
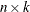 matrix of design points on which the TPSS is to be fit. The
matrix of design points on which the TPSS is to be fit. The 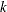 is the number of variables in the spline model. The columns of
is the number of variables in the spline model. The columns of  need to be linearly independent and contain no constant column.
need to be linearly independent and contain no constant column.
- y
-
is the
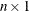 vector of observations.
vector of observations.
- lambda
-
is a optional
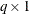 vector that contains
vector that contains 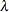 values in
values in 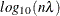 scale. If lambda is not specified (or lambda is specified and
scale. If lambda is not specified (or lambda is specified and 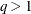 ) the GCV function is used to choose the “best”
) the GCV function is used to choose the “best” 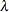 and the returning fitted values are based on the
and the returning fitted values are based on the 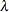 that minimizes the GCV function. If lambda is specified and
that minimizes the GCV function. If lambda is specified and 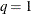 , no minimization of the GCV function is involved and the fitted, coeff and adiag values are all based on the TPSS fit that uses this particular lambda.
, no minimization of the GCV function is involved and the fitted, coeff and adiag values are all based on the TPSS fit that uses this particular lambda.
Aside from the values returned, the TPSPLINE subroutine also prints other useful information such as the number of unique
observations, the dimensions of the null space, the number of parameters in the model, a GCV estimate of ![]() , the smoothing penalty, the residual sum of square, the trace of
, the smoothing penalty, the residual sum of square, the trace of ![]() , an estimate of
, an estimate of ![]() , and the sum of squares for replication.
, and the sum of squares for replication.
No missing values are accepted within the input arguments. Also, you should use caution if you want to specify small lambda values. Since the true ![]() , a very small value for lambda can cause
, a very small value for lambda can cause ![]() to be smaller than the magnitude of machine error and usually the returned gcv values from such a
to be smaller than the magnitude of machine error and usually the returned gcv values from such a ![]() cannot be trusted. Finally, when using TPSPLINE be aware that TPSS is a computationally intensive method. Therefore a large
data set (that is, a large number of unique design points) will take a lot of computer memory and time.
cannot be trusted. Finally, when using TPSPLINE be aware that TPSS is a computationally intensive method. Therefore a large
data set (that is, a large number of unique design points) will take a lot of computer memory and time.
For convenience, the TPSS method is illustrated with a two-dimensional independent variable ![]() . More details can be found in Wahba (1990), or in Bates et al. (1987).
. More details can be found in Wahba (1990), or in Bates et al. (1987).
Assume that the data are from the model
where ![]() are the observations. The function
are the observations. The function ![]() is unknown and you assume that it is reasonably smooth. The error terms
is unknown and you assume that it is reasonably smooth. The error terms ![]() are independent zero-mean random variables.
are independent zero-mean random variables.
You measure the smoothness of ![]() by the integral over the entire plane of the square of the partial derivatives of
by the integral over the entire plane of the square of the partial derivatives of ![]() of total order 2, that is
of total order 2, that is
Using this as a smoothness penalty, the thin-plate smoothing spline estimate ![]() of
of ![]() is the minimizer of
is the minimizer of
Duchon (1976) derived that the minimizer ![]() can be represented as
can be represented as
where ![]() and
and ![]() .
.
Let matrix ![]() have entries
have entries ![]() and matrix
and matrix ![]() have entries
have entries ![]() . Then the minimization problem can be rewritten as finding coefficients
. Then the minimization problem can be rewritten as finding coefficients ![]() and
and ![]() to minimize
to minimize
The final TPSS fits can be viewed as a type of generalized ridge regression estimator. The ![]() is called the smoothing parameter, which controls the balance between the goodness of fit and the smoothness of the final
estimate. The smoothing parameter can be chosen by minimizing the generalized cross validation function (GCV). If you write
is called the smoothing parameter, which controls the balance between the goodness of fit and the smoothness of the final
estimate. The smoothing parameter can be chosen by minimizing the generalized cross validation function (GCV). If you write
and call the ![]() as the “
as the “![]() ” matrix, the GCV function
” matrix, the GCV function ![]() is defined as
is defined as
The returned values from this function call provide the ![]() as fitted, the
as fitted, the ![]() as coeff, and
as coeff, and ![]() as adiag.
as adiag.
To evaluate the TPSS fit ![]() at new data points, you can use the TPSPLNEV call.
at new data points, you can use the TPSPLNEV call.
Suppose ![]() , a
, a ![]() matrix, contains the
matrix, contains the ![]() new data points at which you want to evaluate
new data points at which you want to evaluate ![]() . Let
. Let ![]() and
and ![]() be the
be the ![]() th elements of
th elements of ![]() and
and ![]() respectively. The prediction at new data points
respectively. The prediction at new data points ![]() is
is
Therefore, the ![]() can be easily evaluated by using the coefficient
can be easily evaluated by using the coefficient ![]() obtained from the TPSPLINE call.
obtained from the TPSPLINE call.
An example is given in the documentation for the TPSPLNEV call.