| The HAPLOTYPE Procedure |
Example 8.3 Testing for Linkage Disequilibrium
Again looking at the data from the Lab of Statistical Genetics at Rockefeller University (2001), if you request the test for linkage disequilibrium by specifying the LD option in the PROC HAPLOTYPE statement as follows, the "Test for Allelic Associations" table containing the test statistics is included in the output.
proc haplotype data=ehdata ld;
var m1-m6;
run;
The "Haplotype Frequencies" table (Output 8.3.1) now contains an extra column of the haplotype frequencies under the null hypothesis.
| Haplotype Frequencies | ||||||
|---|---|---|---|---|---|---|
| Number | Haplotype | H0 Freq | H1 Freq | Standard Error |
95% Confidence Limits | |
| 1 | 1-1-1 | 0.08172 | 0.09124 | 0.01353 | 0.06472 | 0.11775 |
| 2 | 1-1-2 | 0.05605 | 0.02124 | 0.00677 | 0.00796 | 0.03452 |
| 3 | 1-1-3 | 0.10006 | 0.11501 | 0.01499 | 0.08563 | 0.14439 |
| 4 | 1-2-1 | 0.09084 | 0.07952 | 0.01271 | 0.05461 | 0.10443 |
| 5 | 1-2-2 | 0.06231 | 0.06726 | 0.01177 | 0.04419 | 0.09032 |
| 6 | 1-2-3 | 0.11122 | 0.12794 | 0.01569 | 0.09718 | 0.15870 |
| 7 | 2-1-1 | 0.08100 | 0.05540 | 0.01075 | 0.03433 | 0.07647 |
| 8 | 2-1-2 | 0.05556 | 0.11690 | 0.01510 | 0.08732 | 0.14649 |
| 9 | 2-1-3 | 0.09918 | 0.07378 | 0.01228 | 0.04971 | 0.09785 |
| 10 | 2-2-1 | 0.09005 | 0.11746 | 0.01513 | 0.08781 | 0.14711 |
| 11 | 2-2-2 | 0.06176 | 0.03028 | 0.00805 | 0.01450 | 0.04606 |
| 12 | 2-2-3 | 0.11025 | 0.10398 | 0.01434 | 0.07587 | 0.13209 |
Note that since the INIT= option was omitted from the PROC HAPLOTYPE statement, the initial haplotype frequencies used in the EM algorithm are identical to the frequencies that appear in the H0 FREQ column in Output 8.3.1. The frequencies in the H1 FREQ column are those calculated from the final iteration of the EM algorithm, and these frequencies’ standard errors and confidence limits are included in the table as well.
Output 8.3.2 displays the log likelihood under the null hypothesis assuming independence among all the loci and the alternative, which allows for associations between markers. The empirical chi-square test statistic of the likelihood ratio test is calculated as 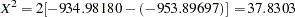 with degrees of freedom
with degrees of freedom  that gives a
that gives a  -value
-value  . The test indicates significant linkage disequilibrium among the three loci, as shown in the online documentation from the Lab of Statistical Genetics at Rockefeller University (2001).
. The test indicates significant linkage disequilibrium among the three loci, as shown in the online documentation from the Lab of Statistical Genetics at Rockefeller University (2001).
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.