| The HAPLOTYPE Procedure |
| Displayed Output |
This section describes the displayed output from PROC HAPLOTYPE. See ODS Table Names for details about how this output interfaces with the Output Delivery System.
Analysis Information
The "Analysis Information" table lists information about the following settings used in PROC HAPLOTYPE for all of the estimation methods:
Loci Used, the loci used to form haplotypes
Number of Individuals
Random Number Seed, the value specified in the SEED= option or generated by the system clock
Haplotype Frequency Cutoff, the value specified in the CUTOFF= option or the default (0)
When EST=EM or EST=STEPEM is specified in the PROC HAPLOTYPE statement, the following information is also included in the table:
Number of Starts, the value specified in the NSTART= option or the default (1)
Convergence Criterion, the value specified in the CONV= option or the default (0.00001)
Iterations Checked for Conv., the value specified in the NLAG= option or the default (1)
Maximum Number of Iterations, the value specified in the MAXITER= option or the default (100)
Number of Iterations Used, as determined by the CONV= or MAXITER= option
Log Likelihood, from the last iteration performed
Initialization Method, the method specified in the INIT= option or "Linkage Equilibrium" by default
Standard Error Method, the method specified in the SE= option or "Binomial" by default
If EST=BAYESIAN is specified in the PROC HAPLOTYPE statement, then these rows are included in the table:
Scaled Mutation Rate, the
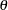 parameter used in the algorithm
parameter used in the algorithm Recorded Iterations, the number of iterations of the algorithm actually recorded, which is

Iteration History
The "Iteration History" table displays the log likelihood and the ratio of change for each iteration of the EM algorithm.
Haplotype Frequencies
The "Haplotype Frequencies" table lists all the possible 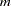 -locus haplotypes in the sample (where
-locus haplotypes in the sample (where 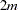 variables are specified in the VAR statement), with an estimate of the haplotype frequency, the standard error of the frequency, and the lower and upper limits of the confidence interval for the frequency based on the confidence level determined by the ALPHA= option of the PROC HAPLOTYPE statement (0.95 by default). When the LD option is specified in the PROC HAPLOTYPE statement and EST=EM or STEPEM, haplotype frequency estimates are calculated both under the null hypothesis of no allelic association by taking the product of allele frequencies, and under the alternative, which allows for associations, using the EM algorithm.
variables are specified in the VAR statement), with an estimate of the haplotype frequency, the standard error of the frequency, and the lower and upper limits of the confidence interval for the frequency based on the confidence level determined by the ALPHA= option of the PROC HAPLOTYPE statement (0.95 by default). When the LD option is specified in the PROC HAPLOTYPE statement and EST=EM or STEPEM, haplotype frequency estimates are calculated both under the null hypothesis of no allelic association by taking the product of allele frequencies, and under the alternative, which allows for associations, using the EM algorithm.
Test for Allelic Associations
The "Test for Allelic Associations" table displays the degrees of freedom and log likelihood calculated using the EM algorithm for the null hypothesis of no association and the alternative hypothesis of associations between markers. The chi-square statistic and its  -value are also shown for the test of these hypotheses.
-value are also shown for the test of these hypotheses.
Test for Marker-Trait Association
The "Test for Marker-Trait Association" table displays the number of observations, degrees of freedom, and log likelihood for both trait values as well as the combined sample when EST=EM or STEPEM. The chi-square test statistic and its corresponding  -value from performing the case-control test, testing the hypothesis of no association between the trait and the marker loci used in PROC HAPLOTYPE, are also given. When the PERMS= option is included in the TRAIT statement, estimates of exact
-value from performing the case-control test, testing the hypothesis of no association between the trait and the marker loci used in PROC HAPLOTYPE, are also given. When the PERMS= option is included in the TRAIT statement, estimates of exact  -values are provided as well.
-values are provided as well.
Tests for Haplotype-Trait Association
The "Tests for Haplotype-Trait Association" table displays statistics from case-control tests performed on each individual haplotype when the TESTALL option is included in the TRAIT statement and EST=EM or STEPEM. A significant  -value indicates that there is an association between the haplotype and the trait. When the PERMS= option is also given in the TRAIT statement, estimates of exact
-value indicates that there is an association between the haplotype and the trait. When the PERMS= option is also given in the TRAIT statement, estimates of exact  -values are provided as well.
-values are provided as well.
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.