| The HAPLOTYPE Procedure |
Overview: HAPLOTYPE Procedure
A haplotype is a combination of alleles at multiple loci on a single chromosome. A pair of haplotypes constitutes the multilocus genotype. Haplotype information has to be inferred because data are usually collected at the genotypic, not haplotype pair, level. For homozygous markers, there is no problem. If one locus has alleles  and
and  , and a second locus has alleles
, and a second locus has alleles  and
and 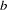 , the observed genotype
, the observed genotype 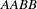 must contain two haplotypes of type
must contain two haplotypes of type 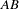 ; genotype
; genotype 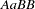 must contain haplotypes
must contain haplotypes 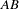 and
and  , and so on. Haplotypes and their frequencies can be obtained directly. When both loci are heterozygous, however, there is ambiguity; a variety of combinations of haplotypes can generate the genotype, and it is not possible to determine directly which two haplotypes constitute any individual genotype. For example, the genotype
, and so on. Haplotypes and their frequencies can be obtained directly. When both loci are heterozygous, however, there is ambiguity; a variety of combinations of haplotypes can generate the genotype, and it is not possible to determine directly which two haplotypes constitute any individual genotype. For example, the genotype  might be of type
might be of type 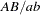 with haplotypes
with haplotypes 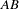 and
and  , or of type
, or of type 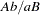 with haplotypes
with haplotypes 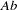 and
and  . The HAPLOTYPE procedure uses the expectation-maximization (EM) algorithm to generate maximum likelihood estimates of haplotype frequencies given a multilocus sample of genetic marker genotypes under the assumption of Hardy-Weinberg equilibrium (HWE). These estimates can then be used to assign the probability that each individual possesses a particular haplotype pair. A Bayesian approach for haplotype frequency estimation is also implemented in PROC HAPLOTYPE.
. The HAPLOTYPE procedure uses the expectation-maximization (EM) algorithm to generate maximum likelihood estimates of haplotype frequencies given a multilocus sample of genetic marker genotypes under the assumption of Hardy-Weinberg equilibrium (HWE). These estimates can then be used to assign the probability that each individual possesses a particular haplotype pair. A Bayesian approach for haplotype frequency estimation is also implemented in PROC HAPLOTYPE.
Estimation of haplotype frequencies is important for several applications in genetic data analysis. One application is determining whether there is linkage disequilibrium (LD), or association, between loci. PROC HAPLOTYPE performs a likelihood ratio test to test the hypothesis of no LD between marker loci. Another application is association testing of disease susceptibility. Since sites that affect disease status are embedded in haplotypes, it has been postulated that the power of case-control studies might be increased by testing for haplotype rather than allele or genotype associations. One reason is that haplotypes might include two or more causative sites whose combined effect is measurable, particularly if they show synergistic interaction. Another reason is that fewer tests need to be performed, although if there are a large number of haplotypes, this advantage is offset by the increased degrees of freedom of each test. PROC HAPLOTYPE can use case-control data to calculate test statistics for the hypothesis of no association between alleles composing the haplotypes and disease status; such tests are carried out across all haplotypes at the loci specified, or for individual haplotypes.
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.